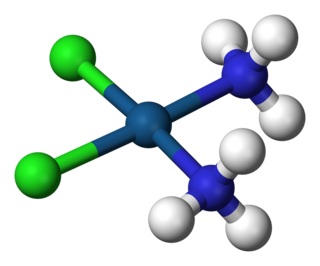
A coordination complex is a chemical compound consisting of a central atom or ion, which is usually metallic and is called the coordination centre, and a surrounding array of bound molecules or ions, that are in turn known as ligands or complexing agents. Many metal-containing compounds, especially those that include transition metals, are coordination complexes.
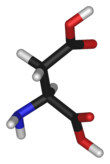
A molecule is a group of two or more atoms held together by attractive forces known as chemical bonds; depending on context, the term may or may not include ions which satisfy this criterion. In quantum physics, organic chemistry, and biochemistry, the distinction from ions is dropped and molecule is often used when referring to polyatomic ions.

A quantum mechanical system or particle that is bound—that is, confined spatially—can only take on certain discrete values of energy, called energy levels. This contrasts with classical particles, which can have any amount of energy. The term is commonly used for the energy levels of the electrons in atoms, ions, or molecules, which are bound by the electric field of the nucleus, but can also refer to energy levels of nuclei or vibrational or rotational energy levels in molecules. The energy spectrum of a system with such discrete energy levels is said to be quantized.

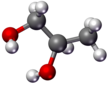
The structural formula of a chemical compound is a graphic representation of the molecular structure, showing how the atoms are possibly arranged in the real three-dimensional space. The chemical bonding within the molecule is also shown, either explicitly or implicitly. Unlike other chemical formula types, which have a limited number of symbols and are capable of only limited descriptive power, structural formulas provide a more complete geometric representation of the molecular structure. For example, many chemical compounds exist in different isomeric forms, which have different enantiomeric structures but the same molecular formula. There are multiple types of ways to draw these structural formulas such as: Lewis Structures, condensed formulas, skeletal formulas, Newman projections, Cyclohexane conformations, Haworth projections, and Fischer projections.
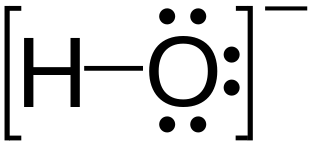
In chemistry, a lone pair refers to a pair of valence electrons that are not shared with another atom in a covalent bond and is sometimes called an unshared pair or non-bonding pair. Lone pairs are found in the outermost electron shell of atoms. They can be identified by using a Lewis structure. Electron pairs are therefore considered lone pairs if two electrons are paired but are not used in chemical bonding. Thus, the number of electrons in lone pairs plus the number of electrons in bonds equals the number of valence electrons around an atom.
Gas electron diffraction (GED) is one of the applications of electron diffraction techniques. The target of this method is the determination of the structure of gaseous molecules, i.e., the geometrical arrangement of the atoms from which a molecule is built up. GED is one of two experimental methods to determine the structure of free molecules, undistorted by intermolecular forces, which are omnipresent in the solid and liquid state. The determination of accurate molecular structures by GED studies is fundamental for an understanding of structural chemistry.

Valence shell electron pair repulsion (VSEPR) theory is a model used in chemistry to predict the geometry of individual molecules from the number of electron pairs surrounding their central atoms. It is also named the Gillespie-Nyholm theory after its two main developers, Ronald Gillespie and Ronald Nyholm.
In chemistry, orbital hybridisation is the concept of mixing atomic orbitals to form new hybrid orbitals suitable for the pairing of electrons to form chemical bonds in valence bond theory. For example, in a carbon atom which forms four single bonds, the valence-shell s orbital combines with three valence-shell p orbitals to form four equivalent sp3 mixtures in a tetrahedral arrangement around the carbon to bond to four different atoms. Hybrid orbitals are useful in the explanation of molecular geometry and atomic bonding properties and are symmetrically disposed in space. Usually hybrid orbitals are formed by mixing atomic orbitals of comparable energies.

In chemistry, a trigonal pyramid is a molecular geometry with one atom at the apex and three atoms at the corners of a trigonal base, resembling a tetrahedron (not to be confused with the tetrahedral geometry). When all three atoms at the corners are identical, the molecule belongs to point group C3v. Some molecules and ions with trigonal pyramidal geometry are the pnictogen hydrides (XH3), xenon trioxide (XeO3), the chlorate ion, ClO−
3, and the sulfite ion, SO2−
3. In organic chemistry, molecules which have a trigonal pyramidal geometry are sometimes described as sp3 hybridized. The AXE method for VSEPR theory states that the classification is AX3E1.

In chemistry, a trigonal bipyramid formation is a molecular geometry with one atom at the center and 5 more atoms at the corners of a triangular bipyramid. This is one geometry for which the bond angles surrounding the central atom are not identical, because there is no geometrical arrangement with five terminal atoms in equivalent positions. Examples of this molecular geometry are phosphorus pentafluoride, and phosphorus pentachloride in the gas phase.

In chemistry, octahedral molecular geometry, also called square bipyramidal, describes the shape of compounds with six atoms or groups of atoms or ligands symmetrically arranged around a central atom, defining the vertices of an octahedron. The octahedron has eight faces, hence the prefix octa. The octahedron is one of the Platonic solids, although octahedral molecules typically have an atom in their centre and no bonds between the ligand atoms. A perfect octahedron belongs to the point group Oh. Examples of octahedral compounds are sulfur hexafluoride SF6 and molybdenum hexacarbonyl Mo(CO)6. The term "octahedral" is used somewhat loosely by chemists, focusing on the geometry of the bonds to the central atom and not considering differences among the ligands themselves. For example, [Co(NH3)6]3+, which is not octahedral in the mathematical sense due to the orientation of the N−H bonds, is referred to as octahedral.
In quantum chemistry, the quantum theory of atoms in molecules (QTAIM), sometimes referred to as atoms in molecules (AIM), is a model of molecular and condensed matter electronic systems in which the principal objects of molecular structure - atoms and bonds - are natural expressions of a system's observable electron density distribution function. An electron density distribution of a molecule is a probability distribution that describes the average manner in which the electronic charge is distributed throughout real space in the attractive field exerted by the nuclei. According to QTAIM, molecular structure is revealed by the stationary points of the electron density together with the gradient paths of the electron density that originate and terminate at these points.
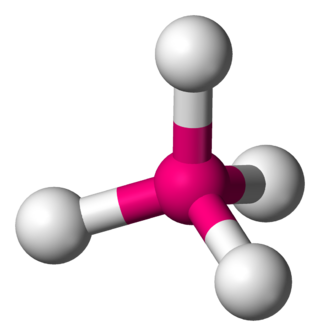
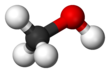
In a tetrahedral molecular geometry, a central atom is located at the center with four substituents that are located at the corners of a tetrahedron. The bond angles are cos−1(−1⁄3) = 109.4712206...° ≈ 109.5° when all four substituents are the same, as in methane as well as its heavier analogues. Methane and other perfectly symmetrical tetrahedral molecules belong to point group Td, but most tetrahedral molecules have lower symmetry. Tetrahedral molecules can be chiral.
In chemistry, Bent's rule describes and explains the relationship between the orbital hybridization and the electronegativities of substituents. The rule was stated by Henry A. Bent as follows:
Atomic s character concentrates in orbitals directed toward electropositive substituents.
A molecular vibration is a periodic motion of the atoms of a molecule relative to each other, such that the center of mass of the molecule remains unchanged. The typical vibrational frequencies range from less than 1013 Hz to approximately 1014 Hz, corresponding to wavenumbers of approximately 300 to 3000 cm−1 and wavelengths of approximately 30 to 3 µm.
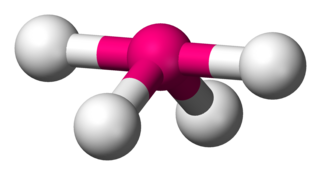
Disphenoidal or seesaw is a type of molecular geometry where there are four bonds to a central atom with overall C2v molecular symmetry. The name "seesaw" comes from the observation that it looks like a playground seesaw. Most commonly, four bonds to a central atom result in tetrahedral or, less commonly, square planar geometry.

The linear molecular geometry describes the geometry around a central atom bonded to two other atoms placed at a bond angle of 180°. Linear organic molecules, such as acetylene, are often described by invoking sp orbital hybridization for their carbon centers.
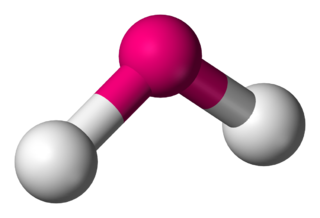
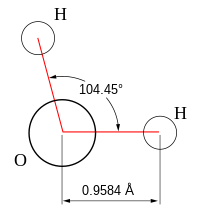
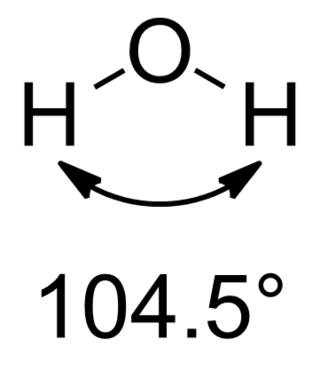
In chemistry, molecules with a non-collinear arrangement of two adjacent bonds have bent molecular geometry, also known as angular or V-shaped. Certain atoms, such as oxygen, will almost always set their two (or more) covalent bonds in non-collinear directions due to their electron configuration. Water (H2O) is an example of a bent molecule, as well as its analogues. The bond angle between the two hydrogen atoms is approximately 104.45°. Nonlinear geometry is commonly observed for other triatomic molecules and ions containing only main group elements, prominent examples being nitrogen dioxide (NO2), sulfur dichloride (SCl2), and methylene (CH2).
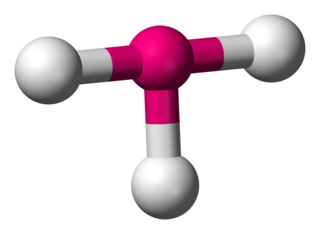
In chemistry, T-shaped molecular geometry describes the structures of some molecules where a central atom has three ligands. Ordinarily, three-coordinated compounds adopt trigonal planar or pyramidal geometries. Examples of T-shaped molecules are the halogen trifluorides, such as ClF3.

In chemistry, isomers are molecules or polyatomic ions with identical molecular formula – that is, same number of atoms of each element – but distinct arrangements of atoms in space. Diamond and graphite are a familiar example; they are isomers of carbon. Isomerism refers to the existence or possibility of isomers.