An internal ribosome entry site, abbreviated IRES, is an RNA element that allows for translation initiation in a cap-independent manner, as part of the greater process of protein synthesis. Initiation of eukaryotic translation nearly always occurs at and is dependent on the 5' cap of mRNA molecules, where the translation initiation complex forms and ribosomes engage the mRNA. IRES elements, however allow ribosomes to engage the mRNA and begin translation independently of the 5' cap.
Bacterial translation is the process by which messenger RNA is translated into proteins in bacteria.
Eukaryotic translation is the biological process by which messenger RNA is translated into proteins in eukaryotes. It consists of four phases: initiation, elongation, termination, and recapping.
Eukaryotic initiation factors (eIFs) are proteins or protein complexes involved in the initiation phase of eukaryotic translation. These proteins help stabilize the formation of ribosomal preinitiation complexes around the start codon and are an important input for post-transcription gene regulation. Several initiation factors form a complex with the small 40S ribosomal subunit and Met-tRNAiMet called the 43S preinitiation complex. Additional factors of the eIF4F complex recruit the 43S PIC to the five-prime cap structure of the mRNA, from which the 43S particle scans 5'-->3' along the mRNA to reach an AUG start codon. Recognition of the start codon by the Met-tRNAiMet promotes gated phosphate and eIF1 release to form the 48S preinitiation complex, followed by large 60S ribosomal subunit recruitment to form the 80S ribosome. There exist many more eukaryotic initiation factors than prokaryotic initiation factors, reflecting the greater biological complexity of eukaryotic translation. There are at least twelve eukaryotic initiation factors, composed of many more polypeptides, and these are described below.
A bacterial initiation factor (IF) is a protein that stabilizes the initiation complex for polypeptide translation.
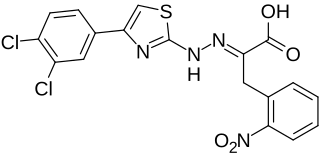
4EGI-1 is a synthetic chemical compound which has been found to interfere with the growth of certain types of cancer cells in vitro. Its mechanism of action involves interruption of the binding of cellular initiation factor proteins involved in the translation of transcribed mRNA at the ribosome. The inhibition of these initiation factors prevents the initiation and translation of many proteins whose functions are essential to the rapid growth and proliferation of cancer cells.
The Hepatitis C virus internal ribosome entry site, or HCV IRES, is an RNA structure within the 5'UTR of the HCV genome that mediates cap-independent translation initiation.

Nuclear cap-binding protein complex is a RNA-binding protein which binds to the 5' cap of pre-mRNA. The cap and nuclear cap-binding protein have many functions in mRNA biogenesis including splicing, 3'-end formation by stabilizing the interaction of the 3'-end processing machinery, nuclear export and protection of the transcripts from nuclease degradation. During mRNA export, the nuclear cap-binding protein complex recruits ribosomes to begin the pioneer round of translation. When RNA is exported to the cytoplasm the nuclear cap-binding protein complex is replaced by cytoplasmic cap binding complex. The nuclear cap-binding complex is a functional heterodimer and composed of Cbc1/Cbc2 in yeast and CBP20/CBP80 in multicellular eukaryotes. Human nuclear cap-binding protein complex shows the large subunit, CBP80 consists of 757 amino acid residues. Its secondary structure contains approximately sixty percent of helical and one percent of beta sheet in the strand. The small subunit, CBP20 has 98 amino acid residues. Its secondary structure contains approximately twenty percent of helical and twenty-four percent of beta sheet in the strand. Human nuclear cap-binding protein complex plays important role in the maturation of pre-mRNA and in uracil-rich small nuclear RNA.

Eukaryotic initiation factor 4A-I is a 46 kDa cytosolic protein that, in humans, is encoded by the EIF4A1 gene, which is located on chromosome 17. It is the most prevalent member of the eIF4A family of ATP-dependant RNA helicases, and plays a critical role in the initiation of cap-dependent eukaryotic protein translation as a component of the eIF4F translation initiation complex. eIF4A1 unwinds the secondary structure of RNA within the 5'-UTR of mRNA, a critical step necessary for the recruitment of the 43S preinitiation complex, and thus the translation of protein in eukaryotes. It was first characterized in 1982 by Grifo, et al., who purified it from rabbit reticulocyte lysate.
Eukaryotic translation initiation factor 1A, X-chromosomal (eIF1A) is a protein that in humans is encoded by the EIF1AX gene. This gene encodes an essential eukaryotic translation initiation factor. The protein is a component of the 43S pre-initiation complex (PIC), which mediates the recruitment of the small 40S ribosomal subunit to the 5' cap of messenger RNAs.

Eukaryotic translation initiation factor 5B is a protein that in humans is encoded by the EIF5B gene.

Eukaryotic translation initiation factor 1 (eIF1) is a protein that in humans is encoded by the EIF1 gene. It is related to yeast SUI1.
Eukaryotic translation initiation factor 4 G (eIF4G) is a protein involved in eukaryotic translation initiation and is a component of the eIF4F cap-binding complex. Orthologs of eIF4G have been studied in multiple species, including humans, yeast, and wheat. However, eIF4G is exclusively found in domain Eukarya, and not in domains Bacteria or Archaea, which do not have capped mRNA. As such, eIF4G structure and function may vary between species, although the human EIF4G1 has been the focus of extensive studies.
Eukaryotic Initiation Factor 2 (eIF2) is an eukaryotic initiation factor. It is required for most forms of eukaryotic translation initiation. eIF2 mediates the binding of tRNAiMet to the ribosome in a GTP-dependent manner. eIF2 is a heterotrimer consisting of an alpha, a beta, and a gamma subunit.
The eukaryotic initiation factor-4A (eIF4A) family consists of 3 closely related proteins EIF4A1, EIF4A2, and EIF4A3. These factors are required for the binding of mRNA to 40S ribosomal subunits. In addition these proteins are helicases that function to unwind double-stranded RNA.
Translational regulation refers to the control of the levels of protein synthesized from its mRNA. This regulation is vastly important to the cellular response to stressors, growth cues, and differentiation. In comparison to transcriptional regulation, it results in much more immediate cellular adjustment through direct regulation of protein concentration. The corresponding mechanisms are primarily targeted on the control of ribosome recruitment on the initiation codon, but can also involve modulation of peptide elongation, termination of protein synthesis, or ribosome biogenesis. While these general concepts are widely conserved, some of the finer details in this sort of regulation have been proven to differ between prokaryotic and eukaryotic organisms.

Eukaryotic initiation factor 3 (eIF3) is a multiprotein complex that functions during the initiation phase of eukaryotic translation. It is essential for most forms of cap-dependent and cap-independent translation initiation. In humans, eIF3 consists of 13 nonidentical subunits (eIF3a-m) with a combined molecular weight of ~800 kDa, making it the largest translation initiation factor. The eIF3 complex is broadly conserved across eukaryotes, but the conservation of individual subunits varies across organisms. For instance, while most mammalian eIF3 complexes are composed of 13 subunits, budding yeast's eIF3 has only six subunits.

Eukaryotic initiation factor 4F (eIF4F) is a heterotrimeric protein complex that binds the 5' cap of messenger RNAs (mRNAs) to promote eukaryotic translation initiation. The eIF4F complex is composed of three non-identical subunits: the DEAD-box RNA helicase eIF4A, the cap-binding protein eIF4E, and the large "scaffold" protein eIF4G. The mammalian eIF4F complex was first described in 1983, and has been a major area of study into the molecular mechanisms of cap-dependent translation initiation ever since.
The 43S preinitiation complex is a ribonucleoprotein complex that exists during an early step of eukaryotic translation initiation. The 43S PIC contains the small ribosomal subunit (40S) bound by the initiation factors eIF1, eIF1A, eIF3, and the eIF2-Met-tRNAiMet-GTP ternary complex (eIF2-TC).
Archaeal initiation factors are proteins that are used during the translation step of protein synthesis in archaea. The principal functions these proteins perform include ribosome RNA/mRNA recognition, delivery of the initiator Met-tRNAiMet, methionine bound tRNAi, to the 40s ribosome, and proofreading of the initiation complex.










