This article relies largely or entirely on a single source .(July 2015) |
The Pierce Protein Assay is a method of protein quantification. It provides quick estimation of the protein amount in a given sample. [1]
This article relies largely or entirely on a single source .(July 2015) |
The Pierce Protein Assay is a method of protein quantification. It provides quick estimation of the protein amount in a given sample. [1]
The assay is separated into three main parts: preparation of the Diluted Albumin (BSA) Standards, preparation of the bicinchoninic acid (BCA) working reagent, and quantification of proteins (using either test tube or microplate procedure).
This method is able to detect as low as 25 μg/ml and up to 2000 μg/ml of protein in a 65 ul sample, using standard protocol. This method may be preferred for samples containing detergents or other reducing agents. This method has a fast detection speed and low protein-to-protein variability in comparison to the BCA or Coomassie (Bradford) Assays. This method has a stable end point.
This method has greater protein-to-protein variability than the BCA Assay.

Reverse transcription polymerase chain reaction (RT-PCR) is an laboratory technique combining reverse transcription of RNA into DNA and amplification of specific DNA targets using polymerase chain reaction (PCR). It is primarily used to measure the amount of a specific RNA. This is achieved by monitoring the amplification reaction using fluorescence, a technique called real-time PCR or quantitative PCR (qPCR). Combined RT-PCR and qPCR are routinely used for analysis of gene expression and quantification of viral RNA in research and clinical settings.
Protein purification is a series of processes intended to isolate one or a few proteins from a complex mixture, usually cells, tissues or whole organisms. Protein purification is vital for the characterization of the function, structure and interactions of the protein of interest. The purification process may separate the protein and non-protein parts of the mixture, and finally separate the desired protein from all other proteins. Separation of one protein from all others is typically the most laborious aspect of protein purification. Separation steps usually exploit differences in protein size, physico-chemical properties, binding affinity and biological activity. The pure result may be termed protein isolate.
In pharmacology, the international unit is a unit of measurement for the amount of a substance; the mass or volume that constitutes one international unit varies based on which substance is being measured, and the variance is based on the biological activity or effect, for the purpose of easier comparison across substances. International units are used to quantify vitamins, hormones, some medications, vaccines, blood products, and similar biologically active substances.
Plate readers, also known as microplate readers or microplate photometers, are instruments which are used to detect biological, chemical or physical events of samples in microtiter plates. They are widely used in research, drug discovery, bioassay validation, quality control and manufacturing processes in the pharmaceutical and biotechnological industry and academic organizations. Sample reactions can be assayed in 1-1536 well format microtiter plates. The most common microplate format used in academic research laboratories or clinical diagnostic laboratories is 96-well with a typical reaction volume between 100 and 200 µL per well. Higher density microplates are typically used for screening applications, when throughput and assay cost per sample become critical parameters, with a typical assay volume between 5 and 50 µL per well. Common detection modes for microplate assays are absorbance, fluorescence intensity, luminescence, time-resolved fluorescence, and fluorescence polarization.
The Bradford protein assay was developed by Marion M. Bradford in 1976. It is a quick and accurate spectroscopic analytical procedure used to measure the concentration of protein in a solution. The reaction is dependent on the amino acid composition of the measured proteins.
Protein methods are the techniques used to study proteins. There are experimental methods for studying proteins. Computational methods typically use computer programs to analyze proteins. However, many experimental methods require computational analysis of the raw data.
The bicinchoninic acid assay, also known as the Smith assay, after its inventor, Paul K. Smith at the Pierce Chemical Company, is a biochemical assay for determining the total concentration of protein in a solution, similar to Lowry protein assay, Bradford protein assay or biuret reagent. The total protein concentration is exhibited by a color change of the sample solution from green to purple in proportion to protein concentration, which can then be measured using colorimetric techniques.

The biuret test, also known as Piotrowski's test, is a chemical test used for detecting the presence of peptide bonds. In the presence of peptides, a copper(II) ion forms mauve-colored coordination complexes in an alkaline solution. Several variants on the test have been developed, such as the BCA test and the Modified Lowry test.
The Kjeldahl method or Kjeldahl digestion (Danish pronunciation: [ˈkʰɛltæːˀl]) in analytical chemistry is a method for the quantitative determination of nitrogen contained in organic substances plus the nitrogen contained in the inorganic compounds ammonia and ammonium (NH3/NH4+). Without modification, other forms of inorganic nitrogen, for instance nitrate, are not included in this measurement. This method was developed by Johan Kjeldahl in 1883.
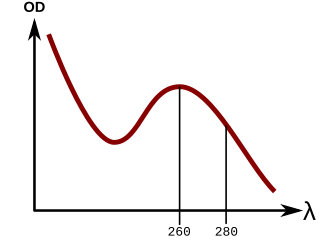
In molecular biology, quantitation of nucleic acids is commonly performed to determine the average concentrations of DNA or RNA present in a mixture, as well as their purity. Reactions that use nucleic acids often require particular amounts and purity for optimum performance. To date, there are two main approaches used by scientists to quantitate, or establish the concentration, of nucleic acids in a solution. These are spectrophotometric quantification and UV fluorescence tagging in presence of a DNA dye.

Quantitative proteomics is an analytical chemistry technique for determining the amount of proteins in a sample. The methods for protein identification are identical to those used in general proteomics, but include quantification as an additional dimension. Rather than just providing lists of proteins identified in a certain sample, quantitative proteomics yields information about the physiological differences between two biological samples. For example, this approach can be used to compare samples from healthy and diseased patients. Quantitative proteomics is mainly performed by two-dimensional gel electrophoresis (2-DE) or mass spectrometry (MS). However, a recent developed method of quantitative dot blot (QDB) analysis is able to measure both the absolute and relative quantity of an individual proteins in the sample in high throughput format, thus open a new direction for proteomic research. In contrast to 2-DE, which requires MS for the downstream protein identification, MS technology can identify and quantify the changes.
Radioimmunoprecipitation assay buffer is a lysis buffer used to lyse cells and tissue for the radio immunoprecipitation assay (RIPA). This buffer is more denaturing than NP-40 or Triton X-100 because it contains the ionic detergents SDS and sodium deoxycholate as active constituents and is particularly useful for disruption of nuclear membranes in the preparation of nuclear extracts. The RIPA buffer gives low background but can denature kinases.
Bioanalysis is a sub-discipline of analytical chemistry covering the quantitative measurement of xenobiotics and biotics in biological systems.
Yessotoxins are a group of lipophilic, sulfur bearing polyether toxins that are related to ciguatoxins. They are produced by a variety of dinoflagellates, most notably Lingulodinium polyedrum and Gonyaulax spinifera.
Virus quantification involves counting the number of viruses in a specific volume to determine the virus concentration. It is utilized in both research and development (R&D) in commercial and academic laboratories as well as production situations where the quantity of virus at various steps is an important variable. For example, the production of viral vaccines, recombinant proteins using viral vectors and viral antigens all require virus quantification to continually adapt and monitor the process in order to optimize production yields and respond to ever changing demands and applications. Examples of specific instances where known viruses need to be quantified include clone screening, multiplicity of infection (MOI) optimization and adaptation of methods to cell culture. This page discusses various techniques currently used to quantify viruses in liquid samples. These methods are separated into two categories, traditional vs. modern methods. Traditional methods are industry-standard methods that have been used for decades but are generally slow and labor-intensive. Modern methods are relatively new commercially available products and kits that greatly reduce quantification time. This is not meant to be an exhaustive review of all potential methods, but rather a representative cross-section of traditional methods and new, commercially available methods. While other published methods may exist for virus quantification, non-commercial methods are not discussed here.
Rockford, IL-based Pierce Chemical Company was founded in 1948 when Dr. Alan Pierce assumed active management of a company known as Midwest Extraction, which focused on extracting chlorophyll from alfalfa. Chlorophyll, the material that lends a green color to plants, was discovered in the 1930s to have therapeutic uses including the treatment of infections and burns and was useful for performing amputations.
Mass spectrometric immunoassay (MSIA) is a rapid method is used to detect and/ or quantify antigens and or antibody analytes. This method uses an analyte affinity isolation to extract targeted molecules and internal standards from biological fluid in preparation for matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF-MS). This method allows for "top down" and "bottom up" analysis. This sensitive method allows for a new and improved process for detecting multiple antigens and antibodies in a single assay. This assay is also capable of distinguishing mass shifted forms of the same molecule via a panantibody, as well as distinguish point mutations in proteins. Each specific form is detected uniquely based on their characteristic molecular mass. MSIA has dual specificity because of the antibody-antigen reaction coupled with the power of a mass spectrometer.
In the field of cellular biology, single-cell analysis is the study of genomics, transcriptomics, proteomics and metabolomics at the single cell level. Due to the heterogeneity seen in both eukaryotic and prokaryotic cell populations, analyzing a single cell makes it possible to discover mechanisms not seen when studying a bulk population of cells. Technologies such as fluorescence-activated cell sorting (FACS) or digital dielectrophoretic sorting (DEPArray), allow the precise isolation of selected single cells from complex samples, while high throughput single cell partitioning technologies, enable the simultaneous molecular analysis of hundreds or thousands single unsorted cells; this is particularly useful for the analysis of transcriptome variation in genotypically identical cells, allowing the definition of otherwise undetectable cell subtypes. The development of new technologies is increasing our ability to analyze the genome, and transcriptome, of single cells, as well as to quantify their proteome and metabolome. New developments in mass spectrometry techniques have become important analytical tools for proteomic and metabolomic analysis of single cells. In situ sequencing and fluorescence in situ hybridization (FISH) do not require that cells be isolated and are increasingly being used for analysis of tissues.
Normalization of Western blot data is an analytical step that is performed to compare the relative abundance of a specific protein across the lanes of a blot or gel under diverse experimental treatments, or across tissues or developmental stages. The overall goal of normalization is to minimize effects arising from variations in experimental errors, such as inconsistent sample preparation, unequal sample loading across gel lanes, or uneven protein transfer, which can compromise the conclusions that can be obtained from Western blot data. Currently, there are two methods for normalizing Western blot data: (i) housekeeping protein normalization and (ii) total protein normalization.
Stable isotope standards and capture by anti-peptide antibodies (SISCAPA) is a mass spectrometry method for measuring the amount of a protein in a biological sample.