Related Research Articles

Enzymes are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called enzymology and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties.

Thiamine, also known as thiamin and vitamin B1, is a vitamin, an essential micronutrient for humans and animals. It is found in food and commercially synthesized to be a dietary supplement or medication. Phosphorylated forms of thiamine are required for some metabolic reactions, including the breakdown of glucose and amino acids.

A hemeprotein, or heme protein, is a protein that contains a heme prosthetic group. They are a very large class of metalloproteins. The heme group confers functionality, which can include oxygen carrying, oxygen reduction, electron transfer, and other processes. Heme is bound to the protein either covalently or noncovalently or both.

Heme, or haem, is a precursor to hemoglobin, which is necessary to bind oxygen in the bloodstream. Heme is biosynthesized in both the bone marrow and the liver.

A biomolecule or biological molecule is a loosely used term for molecules present in organisms that are essential to one or more typically biological processes, such as cell division, morphogenesis, or development. Biomolecules include large macromolecules such as proteins, carbohydrates, lipids, and nucleic acids, as well as small molecules such as primary metabolites, secondary metabolites and natural products. A more general name for this class of material is biological materials. Biomolecules are an important element of living organisms, those biomolecules are often endogenous, produced within the organism but organisms usually need exogenous biomolecules, for example certain nutrients, to survive.

A cofactor is a non-protein chemical compound or metallic ion that is required for an enzyme's role as a catalyst. Cofactors can be considered "helper molecules" that assist in biochemical transformations. The rates at which these happen are characterized in an area of study called enzyme kinetics. Cofactors typically differ from ligands in that they often derive their function by remaining bound.

Pyrroloquinoline quinone (PQQ), also called methoxatin, is a redox cofactor and antioxidant. Produced by bacteria, it is found in soil and foods such as kiwifruit, as well as human breast milk. Enzymes using PQQ as a redox cofactor are called quinoproteins and play a variety of redox roles. Quinoprotein glucose dehydrogenase is used as a glucose sensor in bacteria. PQQ stimulates growth in bacteria. Eukaryote targets, including mammalian lactate dehydrogenase, are of more interest to health. It is suggested that PQQ taken as a dietary supplement could promote mitochondrial biogenesis via this pathway as well as PGC-1α.
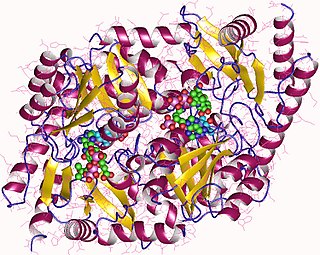
Aminolevulinic acid synthase (ALA synthase, ALAS, or delta-aminolevulinic acid synthase) is an enzyme (EC 2.3.1.37) that catalyzes the synthesis of δ-aminolevulinic acid (ALA) the first common precursor in the biosynthesis of all tetrapyrroles such as hemes, cobalamins and chlorophylls. The reaction is as follows:

Pyridoxal phosphate (PLP, pyridoxal 5'-phosphate, P5P), the active form of vitamin B6, is a coenzyme in a variety of enzymatic reactions. The International Union of Biochemistry and Molecular Biology has catalogued more than 140 PLP-dependent activities, corresponding to ~4% of all classified activities. The versatility of PLP arises from its ability to covalently bind the substrate, and then to act as an electrophilic catalyst, thereby stabilizing different types of carbanionic reaction intermediates.

Succinate dehydrogenase (SDH) or succinate-coenzyme Q reductase (SQR) or respiratory complex II is an enzyme complex, found in many bacterial cells and in the inner mitochondrial membrane of eukaryotes. It is the only enzyme that participates in both the citric acid cycle and the electron transport chain. Histochemical analysis showing high succinate dehydrogenase in muscle demonstrates high mitochondrial content and high oxidative potential.

In biochemistry, flavin adenine dinucleotide (FAD) is a redox-active coenzyme associated with various proteins, which is involved with several enzymatic reactions in metabolism. A flavoprotein is a protein that contains a flavin group, which may be in the form of FAD or flavin mononucleotide (FMN). Many flavoproteins are known: components of the succinate dehydrogenase complex, α-ketoglutarate dehydrogenase, and a component of the pyruvate dehydrogenase complex.

Transketolase is an enzyme that, in humans, is encoded by the TKT gene. It participates in both the pentose phosphate pathway in all organisms and the Calvin cycle of photosynthesis. Transketolase catalyzes two important reactions, which operate in opposite directions in these two pathways. In the first reaction of the non-oxidative pentose phosphate pathway, the cofactor thiamine diphosphate accepts a 2-carbon fragment from a 5-carbon ketose (D-xylulose-5-P), then transfers this fragment to a 5-carbon aldose (D-ribose-5-P) to form a 7-carbon ketose (sedoheptulose-7-P). The abstraction of two carbons from D-xylulose-5-P yields the 3-carbon aldose glyceraldehyde-3-P. In the Calvin cycle, transketolase catalyzes the reverse reaction, the conversion of sedoheptulose-7-P and glyceraldehyde-3-P to pentoses, the aldose D-ribose-5-P and the ketose D-xylulose-5-P.
A holoprotein or conjugated protein is an apoprotein combined with its prosthetic group.
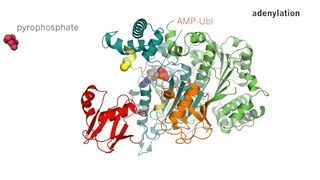
Enzyme catalysis is the increase in the rate of a process by a biological molecule, an "enzyme". Most enzymes are proteins, and most such processes are chemical reactions. Within the enzyme, generally catalysis occurs at a localized site, called the active site.
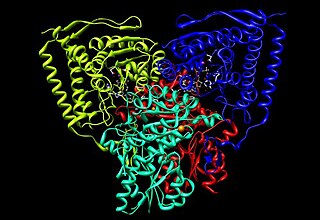
Pyruvate dehydrogenase is an enzyme that catalyzes the reaction of pyruvate and a lipoamide to give the acetylated dihydrolipoamide and carbon dioxide. The conversion requires the coenzyme thiamine pyrophosphate.

Heme C is an important kind of heme.

Pyridoxine 5′-phosphate oxidase is an enzyme, encoded by the PNPO gene, that catalyzes several reactions in the vitamin B6 metabolism pathway. Pyridoxine 5′-phosphate oxidase catalyzes the final, rate-limiting step in vitamin B6 metabolism, the biosynthesis of pyridoxal 5′-phosphate, the biologically active form of vitamin B6 which acts as an essential cofactor. Pyridoxine 5′-phosphate oxidase is a member of the enzyme class oxidases, or more specifically, oxidoreductases. These enzymes catalyze a simultaneous oxidation-reduction reaction. The substrate oxidase enzymes is hydroxlyated by one oxygen atom of molecular oxygen. Concurrently, the other oxygen atom is reduced to water. Even though molecular oxygen is the electron acceptor in these enzymes' reactions, they are unique because oxygen does not appear in the oxidized product.
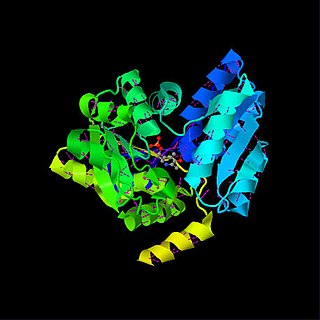
Serine dehydratase or L-serine ammonia lyase (SDH) is in the β-family of pyridoxal phosphate-dependent (PLP) enzymes. SDH is found widely in nature, but its structural and properties vary among species. SDH is found in yeast, bacteria, and the cytoplasm of mammalian hepatocytes. SDH catalyzes is the deamination of L-serine to yield pyruvate, with the release of ammonia.
In molecular biology, the Cys/Met metabolism PLP-dependent enzyme family is a family of proteins including enzymes involved in cysteine and methionine metabolism which use PLP (pyridoxal-5'-phosphate) as a cofactor.
Ruma Banerjee is a professor of enzymology and biological chemistry at the University of Michigan Medical School. She is an experimentalist whose research has focused on unusual cofactors in enzymology.
References
- ↑ de Bolster, M.W.G. (1997). "Glossary of Terms Used in Bioinorganic Chemistry: Prosthetic groups". International Union of Pure and Applied Chemistry. Archived from the original on 2012-11-28. Retrieved 2007-10-30.
- ↑ Metzler DE (2001) Biochemistry. The chemical reactions of living cells, 2nd edition, Harcourt, San Diego.
- ↑ Nelson DL and Cox M.M (2000) Lehninger, Principles of Biochemistry, 3rd edition, Worth Publishers, New York
- ↑ Campbell MK and Farrell SO (2009) Biochemistry, 6th edition, Thomson Brooks/Cole, Belmont, California
- 1 2 Joosten V, van Berkel WJ (2007). "Flavoenzymes". Curr Opin Chem Biol. 11 (2): 195–202. doi:10.1016/j.cbpa.2007.01.010. PMID 17275397.
- ↑ Salisbury SA, Forrest HS, Cruse WB, Kennard O (1979). "A novel coenzyme from bacterial primary alcohol dehydrogenases". Nature. 280 (5725): 843–4. Bibcode:1979Natur.280..843S. doi:10.1038/280843a0. PMID 471057. S2CID 3094647.
{{cite journal}}: CS1 maint: multiple names: authors list (link) PMID 471057 - ↑ Eliot AC, Kirsch JF (2004). "Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations". Annu. Rev. Biochem. 73: 383–415. doi:10.1146/annurev.biochem.73.011303.074021. PMID 15189147.
- ↑ Jitrapakdee S, Wallace JC (2003). "The biotin enzyme family: conserved structural motifs and domain rearrangements". Curr. Protein Pept. Sci. 4 (3): 217–29. doi:10.2174/1389203033487199. PMID 12769720.
- ↑ Banerjee R, Ragsdale SW (2003). "The many faces of vitamin B12: catalysis by cobalamin-dependent enzymes". Annu. Rev. Biochem. 72: 209–47. doi:10.1146/annurev.biochem.72.121801.161828. PMID 14527323. S2CID 37393683.
- ↑ Frank RA, Leeper FJ, Luisi BF (2007). "Structure, mechanism and catalytic duality of thiamine-dependent enzymes". Cell. Mol. Life Sci. 64 (7–8): 892–905. doi:10.1007/s00018-007-6423-5. PMID 17429582. S2CID 20415735.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Wijayanti N, Katz N, Immenschuh S (2004). "Biology of heme in health and disease". Curr. Med. Chem. 11 (8): 981–6. doi:10.2174/0929867043455521. PMID 15078160.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Mendel RR, Hänsch R (2002). "Molybdoenzymes and molybdenum cofactor in plants". J. Exp. Bot. 53 (375): 1689–98. doi: 10.1093/jxb/erf038 . PMID 12147719.
- ↑ Mendel RR, Bittner F (2006). "Cell biology of molybdenum". Biochim. Biophys. Acta. 1763 (7): 621–35. doi:10.1016/j.bbamcr.2006.03.013. PMID 16784786.
- ↑ Bustamante J, Lodge JK, Marcocci L, Tritschler HJ, Packer L, Rihn BH (1998). "Alpha-lipoic acid in liver metabolism and disease". Free Radic. Biol. Med. 24 (6): 1023–39. doi:10.1016/S0891-5849(97)00371-7. PMID 9607614.
{{cite journal}}: CS1 maint: multiple names: authors list (link)