
Nicotinamide adenine dinucleotide (NAD) is a coenzyme central to metabolism. Found in all living cells, NAD is called a dinucleotide because it consists of two nucleotides joined through their phosphate groups. One nucleotide contains an adenine nucleobase and the other nicotinamide. NAD exists in two forms: an oxidized and reduced form, abbreviated as NAD+ and NADH (H for hydrogen), respectively.

Poly (ADP-ribose) polymerase (PARP) is a family of proteins involved in a number of cellular processes such as DNA repair, genomic stability, and programmed cell death.

ADP-ribosylation is the addition of one or more ADP-ribose moieties to a protein. It is a reversible post-translational modification that is involved in many cellular processes, including cell signaling, DNA repair, gene regulation and apoptosis. Improper ADP-ribosylation has been implicated in some forms of cancer. It is also the basis for the toxicity of bacterial compounds such as cholera toxin, diphtheria toxin, and others.

Poly [ADP-ribose] polymerase 1 (PARP-1) also known as NAD+ ADP-ribosyltransferase 1 or poly[ADP-ribose] synthase 1 is an enzyme that in humans is encoded by the PARP1 gene. It is the most abundant of the PARP family of enzymes, accounting for 90% of the NAD+ used by the family. PARP1 is mostly present in cell nucleus, but cytosolic fraction of this protein was also reported.

ADP-ribose diphosphatase (EC 3.6.1.13) is an enzyme that catalyzes a hydrolysis reaction in which water nucleophilically attacks ADP-ribose to produce AMP and D-ribose 5-phosphate. Enzyme hydrolysis occurs by the breakage of a phosphoanhydride bond and is dependent on Mg2+ ions that are held in complex by the enzyme.
In enzymology, an ADP-ribosyl-[dinitrogen reductase] hydrolase is an enzyme that catalyzes the chemical reaction
In enzymology, a NAD+ glycohydrolase (EC 3.2.2.5) is an enzyme that catalyzes the chemical reaction

In enzymology, a ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase (EC 3.2.2.6) is a bifunctional enzyme that catalyzes the chemical reaction
In enzymology, a NAD(P)+-protein-arginine ADP-ribosyltransferase (EC 2.4.2.31) is an enzyme that catalyzes the chemical reaction using nicotinamide adenine dinucleotide
In enzymology, a NAD+-dinitrogen-reductase ADP-D-ribosyltransferase (EC 2.4.2.37) is an enzyme that catalyzes the chemical reaction
Tankyrase, also known as tankyrase 1, is an enzyme that in humans is encoded by the TNKS gene. It inhibits the binding of TERF1 to telomeric DNA. Tankyrase attracts substantial interest in cancer research through its interaction with AXIN1 and AXIN2, which are negative regulators of pro-oncogenic β-catenin signaling. Importantly, activity in the β-catenin destruction complex can be increased by tankyrase inhibitors and thus such inhibitors are a potential therapeutic option to reduce the growth of β-catenin-dependent cancers.

Poly [ADP-ribose] polymerase 4 is an enzyme that in humans is encoded by the PARP4 gene.
Poly [ADP-ribose] polymerase 3 is an enzyme that in humans is encoded by the PARP3 gene.

Poly [ADP-ribose] polymerase 2 is an enzyme that in humans is encoded by the PARP2 gene. It is one of the PARP family of enzymes.
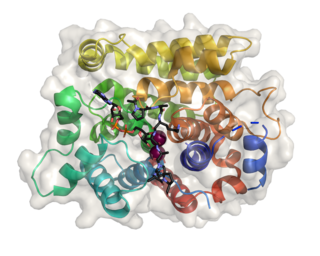
(ADP-ribosyl)hydrolase 3 (ARH3) is an enzyme that in humans is encoded by the ADPRHL2 gene (also called ADPRS). This enzyme reverses the proteins’ post-translational addition of ADP-ribose to serine residues as part of the DNA damage response The enzyme is also known to cleave poly(ADP-ribose) polymers, 1''-O-acetyl-ADP-ribose and alpha-NAD+
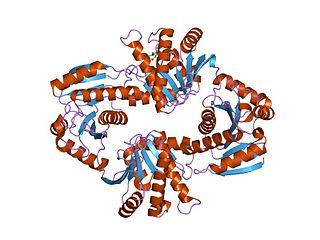
In molecular biology, the Macro domain or A1pp domain is a module of about 180 amino acids which can bind ADP-ribose, an NAD metabolite, or related ligands. Binding to ADP-ribose can be either covalent or non-covalent: in certain cases it is believed to bind non-covalently, while in other cases it appears to bind both non-covalently through a zinc finger motif, and covalently through a separate region of the protein.
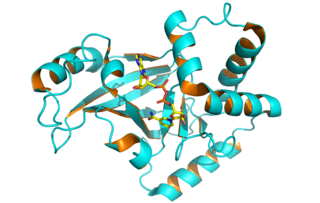
Clostridium botulinum C3 exoenzyme is a toxin that causes the addition of one or more ADP-ribose moieties to Rho-like proteins. Many bacterial toxins nucleotide-binding modify by ADP-ribosylation proteins involved in essential cell functions, leading to their toxic effects.

Poly (ADP-ribose) glycohydrolase is an enzyme that in humans is encoded by the PARG gene.

(ADP-ribosyl)hydrolase 1, also termed [Protein ADP-ribosylarginine] hydrolase and protein-Nω-(ADP-D-ribosyl)-L-arginine ADP-ribosylhydrolase (EC 3.2.2.19), is an enzyme that in humans is encoded by the ADPRH gene. This enzyme is a specific mono(ADP-ribosyl)hydrolase that catalyses the removal of an ADP-ribosyl modification from target arginine residues of protein substrates. The chemical reactions can formally be described as follows:

(ADP-ribosyl)hydrolase 2 (ARH2) is a protein that in humans is encoded by the ADPRHL1 gene.
















