Related Research Articles

Meiosis is a special type of cell division of germ cells and apicomplexans in sexually-reproducing organisms that produces the gametes, such as sperm or egg cells. It involves two rounds of division that ultimately result in four cells with only one copy of each chromosome (haploid). Additionally, prior to the division, genetic material from the paternal and maternal copies of each chromosome is crossed over, creating new combinations of code on each chromosome. Later on, during fertilisation, the haploid cells produced by meiosis from a male and a female will fuse to create a cell with two copies of each chromosome again, the zygote.

Chromosomal crossover, or crossing over, is the exchange of genetic material during sexual reproduction between two homologous chromosomes' non-sister chromatids that results in recombinant chromosomes. It is one of the final phases of genetic recombination, which occurs in the pachytene stage of prophase I of meiosis during a process called synapsis. Synapsis begins before the synaptonemal complex develops and is not completed until near the end of prophase I. Crossover usually occurs when matching regions on matching chromosomes break and then reconnect to the other chromosome.
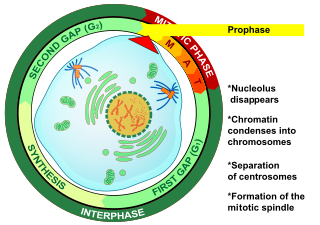
Prophase is the first stage of cell division in both mitosis and meiosis. Beginning after interphase, DNA has already been replicated when the cell enters prophase. The main occurrences in prophase are the condensation of the chromatin reticulum and the disappearance of the nucleolus.

A couple of homologous chromosomes, or homologs, are a set of one maternal and one paternal chromosome that pair up with each other inside a cell during fertilization. Homologs have the same genes in the same loci, where they provide points along each chromosome that enable a pair of chromosomes to align correctly with each other before separating during meiosis. This is the basis for Mendelian inheritance, which characterizes inheritance patterns of genetic material from an organism to its offspring parent developmental cell at the given time and area.

Nondisjunction is the failure of homologous chromosomes or sister chromatids to separate properly during cell division (mitosis/meiosis). There are three forms of nondisjunction: failure of a pair of homologous chromosomes to separate in meiosis I, failure of sister chromatids to separate during meiosis II, and failure of sister chromatids to separate during mitosis. Nondisjunction results in daughter cells with abnormal chromosome numbers (aneuploidy).

Synapsis is the pairing of two chromosomes that occurs during meiosis. It allows matching-up of homologous pairs prior to their segregation, and possible chromosomal crossover between them. Synapsis takes place during prophase I of meiosis. When homologous chromosomes synapse, their ends are first attached to the nuclear envelope. These end-membrane complexes then migrate, assisted by the extranuclear cytoskeleton, until matching ends have been paired. Then the intervening regions of the chromosome are brought together, and may be connected by a protein-RNA complex called the synaptonemal complex. During synapsis, autosomes are held together by the synaptonemal complex along their whole length, whereas for sex chromosomes, this only takes place at one end of each chromosome.
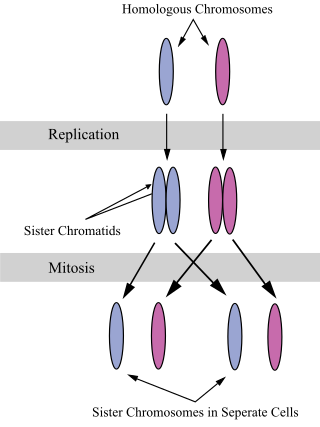
A sister chromatid refers to the identical copies (chromatids) formed by the DNA replication of a chromosome, with both copies joined together by a common centromere. In other words, a sister chromatid may also be said to be 'one-half' of the duplicated chromosome. A pair of sister chromatids is called a dyad. A full set of sister chromatids is created during the synthesis (S) phase of interphase, when all the chromosomes in a cell are replicated. The two sister chromatids are separated from each other into two different cells during mitosis or during the second division of meiosis.
Mitotic recombination is a type of genetic recombination that may occur in somatic cells during their preparation for mitosis in both sexual and asexual organisms. In asexual organisms, the study of mitotic recombination is one way to understand genetic linkage because it is the only source of recombination within an individual. Additionally, mitotic recombination can result in the expression of recessive alleles in an otherwise heterozygous individual. This expression has important implications for the study of tumorigenesis and lethal recessive alleles. Mitotic homologous recombination occurs mainly between sister chromatids subsequent to replication. Inter-sister homologous recombination is ordinarily genetically silent. During mitosis the incidence of recombination between non-sister homologous chromatids is only about 1% of that between sister chromatids.

Sister chromatid exchange (SCE) is the exchange of genetic material between two identical sister chromatids.

A bivalent is one pair of chromosomes in a tetrad. A tetrad is the association of a pair of homologous chromosomes physically held together by at least one DNA crossover. This physical attachment allows for alignment and segregation of the homologous chromosomes in the first meiotic division. In most organisms, each replicated chromosome elicits formation of DNA double-strand breaks during the leptotene phase. These breaks are repaired by homologous recombination, that uses the homologous chromosome as a template for repair. The search for the homologous target, helped by numerous proteins collectively referred as the synaptonemal complex, cause the two homologs to pair, between the leptotene and the pachytene phases of meiosis I.
Chromosome segregation is the process in eukaryotes by which two sister chromatids formed as a consequence of DNA replication, or paired homologous chromosomes, separate from each other and migrate to opposite poles of the nucleus. This segregation process occurs during both mitosis and meiosis. Chromosome segregation also occurs in prokaryotes. However, in contrast to eukaryotic chromosome segregation, replication and segregation are not temporally separated. Instead segregation occurs progressively following replication.

Meiotic recombination protein REC8 homolog is a protein that in humans is encoded by the REC8 gene.
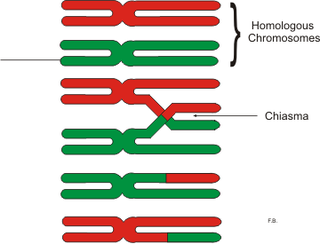
In genetics, a chiasma is the point of contact, the physical link, between two (non-sister) chromatids belonging to homologous chromosomes. At a given chiasma, an exchange of genetic material can occur between both chromatids, what is called a chromosomal crossover, but this is much more frequent during meiosis than mitosis. In meiosis, absence of a chiasma generally results in improper chromosomal segregation and aneuploidy.

Goniaea australasiae is a species of grasshopper in the family Acrididae.

The meiotic recombination checkpoint monitors meiotic recombination during meiosis, and blocks the entry into metaphase I if recombination is not efficiently processed.
The origin and function of meiosis are currently not well understood scientifically, and would provide fundamental insight into the evolution of sexual reproduction in eukaryotes. There is no current consensus among biologists on the questions of how sex in eukaryotes arose in evolution, what basic function sexual reproduction serves, and why it is maintained, given the basic two-fold cost of sex. It is clear that it evolved over 1.2 billion years ago, and that almost all species which are descendants of the original sexually reproducing species are still sexual reproducers, including plants, fungi, and animals.

Neocentromeres are new centromeres that form at a place on the chromosome that is usually not centromeric. They typically arise due to disruption of the normal centromere. These neocentromeres should not be confused with “knobs”, which were also described as “neocentromeres” in maize in the 1950s. Unlike most normal centromeres, neocentromeres do not contain satellite sequences that are highly repetitive but instead consist of unique sequences. Despite this, most neocentromeres are still able to carry out the functions of normal centromeres in regulating chromosome segregation and inheritance. This raises many questions on what is necessary versus what is sufficient for constituting a centromere.
Holocentric chromosomes are chromosomes that possess multiple kinetochores along their length rather than the single centromere typical of other chromosomes. They were first described in cytogenetic experiments in 1935. Since this first observation, the term holocentric chromosome has referred to chromosomes that: i) lack the primary constriction corresponding to the centromere observed in monocentric chromosomes; and ii) possess multiple kinetochores dispersed along the entire chromosomal axis, such that microtubules bind to the chromosome along its entire length and move broadside to the pole from the metaphase plate. Holocentric chromosomes are also termed holokinetic, because, during cell division, the sister chromatids move apart in parallel and do not form the classical V-shaped figures typical of monocentric chromosomes.
Abby F. Dernburg is a professor of Cell and Developmental Biology at the University of California, Berkeley, an Investigator of the Howard Hughes Medical Institute, and a Faculty Senior Scientist at Lawrence Berkeley National Laboratory.
Ovum quality is the measure of the ability of an oocyte to achieve successful fertilisation. The quality is determined by the maturity of the oocyte and the cells that it comprises, which are susceptible to various factors which impact quality and thus reproductive success. This is of significance as an embryo's development is more heavily reliant on the oocyte in comparison to the sperm.
References
- 1 2 King, Robert C. (2013). A dictionary of genetics. Pamela Khipple Mulligan, William D. Stansfield (8th ed.). New York: Oxford University Press. ISBN 978-0-19-937686-5. OCLC 871046520.
- 1 2 3 4 5 6 Kurdzo, Emily L.; Dawson, Dean S. (July 2015). "Centromere pairing – tethering partner chromosomes in meiosis I". The FEBS Journal. 282 (13): 2458–2470. doi:10.1111/febs.13280. ISSN 1742-464X. PMC 4490064 . PMID 25817724.
- 1 2 Kurdzo, Emily L.; Chuong, Hoa H.; Evatt, Jared M.; Dawson, Dean S. (2018-08-09). Sullivan, Beth A. (ed.). "A ZIP1 separation-of-function allele reveals that centromere pairing drives meiotic segregation of achiasmate chromosomes in budding yeast". PLOS Genetics. 14 (8): e1007513. doi: 10.1371/journal.pgen.1007513 . ISSN 1553-7404. PMC 6103513 . PMID 30091974.
- 1 2 3 Dedukh, Dmitrij; da Cruz, Irene; Kneitz, Susanne; Marta, Anatolie; Ormanns, Jenny; Tichopád, Tomáš; Lu, Yuan; Alsheimer, Manfred; Janko, Karel; Schartl, Manfred (December 2022). "Achiasmatic meiosis in the unisexual Amazon molly, Poecilia formosa". Chromosome Research. 30 (4): 443–457. doi:10.1007/s10577-022-09708-2. ISSN 0967-3849. PMC 9771850 . PMID 36459298.
- 1 2 3 Wolf, Klaus Werner (February 1994). "How meiotic cells deal with non-exchange chromosomes". BioEssays. 16 (2): 107–114. doi:10.1002/bies.950160207. ISSN 0265-9247. PMID 8147841. S2CID 32544159.
- 1 2 3 Bosco, Giovanni (2009-02-06). Copenhaver, Gregory P. (ed.). "When Segregation Hangs by a Thread". PLOS Genetics. 5 (2): e1000371. doi: 10.1371/journal.pgen.1000371 . ISSN 1553-7404. PMC 2631148 . PMID 19197362.
- 1 2 Hughes, Stacie E.; Gilliland, William D.; Cotitta, Jeffrey L.; Takeo, Satomi; Collins, Kim A.; Hawley, R. Scott (2009-01-23). Copenhaver, Gregory P. (ed.). "Heterochromatic Threads Connect Oscillating Chromosomes during Prometaphase I in Drosophila Oocytes". PLOS Genetics. 5 (1): e1000348. doi: 10.1371/journal.pgen.1000348 . ISSN 1553-7404. PMC 2615114 . PMID 19165317.
- 1 2 Papaioannou, Ioannis A.; Dutreux, Fabien; Peltier, France A.; Maekawa, Hiromi; Delhomme, Nicolas; Bardhan, Amit; Friedrich, Anne; Schacherer, Joseph; Knop, Michael (December 2021). "Sex without crossing over in the yeast Saccharomycodes ludwigii". Genome Biology. 22 (1): 303. doi: 10.1186/s13059-021-02521-w . ISSN 1474-760X. PMC 8567612 . PMID 34732243.
- 1 2 Tsai, Jui-He; Yan, Rihui; McKee, Bruce D. (August 2011). "Homolog pairing and sister chromatid cohesion in heterochromatin in Drosophila male meiosis I". Chromosoma. 120 (4): 335–351. doi:10.1007/s00412-011-0314-0. ISSN 0009-5915. PMID 21384262. S2CID 1673060.
- ↑ Grozeva, Snejana; Nokkala, Seppo; Simov, Nikolay (2009-12-29). "Chiasmate male meiosis in six species of water bugs from infraorders Nepomorpha and Gerromorpha (Insecta: Heteroptera)". Comparative Cytogenetics. 3 (2): 125–130. doi: 10.3897/compcytogen.v3i2.19 . ISSN 1993-078X.
- ↑ Stoianova, Desislava; Grozeva, Snejana; Simov, Nikolay; Kuznetsova, Valentina (2015-11-19). "Achiasmate male meiosis in two Cymatia species (Hemiptera, Heteroptera, Corixidae)". ZooKeys (538): 95–104. doi: 10.3897/zookeys.538.6722 . ISSN 1313-2970. PMC 4722919 . PMID 26807038.
- ↑ Eyster, Craig; Chuong, Hoa H.; Lee, Chih-Ying; Pezza, Roberto J.; Dawson, Dean (September 2019). "The pericentromeric heterochromatin of homologous chromosomes remains associated after centromere pairing dissolves in mouse spermatocyte meiosis". Chromosoma. 128 (3): 355–367. doi:10.1007/s00412-019-00708-6. ISSN 0009-5915. PMC 6823320 . PMID 31165256.