Preliminaries
Recall that a tree (or a forest) is irreductible when it lacks any internal node of degree 2. In the case of a rooted tree (or a rooted forest), the root(s) are of course allowed to have degree 2, since they are not internal nodes. Any tree (or forest) can be made irreductible by applying a sequence of edge contractions.
An irreductible (rooted or unrooted) tree T whose leaves are bijectively labeled by elements of a set X is called a (rooted or unrooted) X-tree. Such a X-tree usually model a phylogenetic tree, where the elements of X (the taxon set) could represent species, individual organisms, DNA sequences, or other biological objects.
Two X-trees T1 and T2 are said to be isomorphic when there exists a graph isomorphism between them which preserves the leaf labels. In the case of rooted X-trees, the isomorphism must also preserves the root.
Given a X-tree T and a taxon subset Y ⊆ X, the minimal subtree of T that connects all leaves in Y is denoted by T(Y). When T is rooted, then T(Y) is also rooted, with its root being the node closest to the original root of T. This T(Y) subtree needs not be a Y-tree, because it might not be irreductible. We therefore further define the restricted subtree T|Y, which is obtained from T(Y) by suppressing all internal nodes of degree 2, yielding a proper Y-tree.
Agreement forests
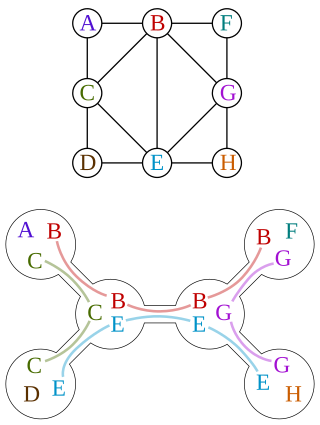
An agreement forest for two unrootedX-trees T1 and T2 is a partition {X1, X2, ..., Xk} of the taxon set X satisfying the following conditions:
- T1|Xi and T2|Xi are isomorphic for every i ∈ {1,2,...,k} and
- the subtrees in { T1(Xi) : i ∈ {1,2,...,k} } and { T2(Xi) : i ∈ {1,2,...,k} } are vertex-disjoint subtrees of T1 and T2, respectively.
The set partition {X1, X2, ..., Xk} is identified with the forest of restricted subtrees F = {T|X1, T|X2, ..., T|Xk}, with either T=T1 or T=T2 (the choice of it begin irrelevant because of condition 1). Therefore, an agreement forest can either be seen as a partition of the taxon set X or as a forest (in the classical graph-theoretic sense) of restricted subtrees.
The size of an agreement forest is simply its number of components. Intuitively, an agreement forest of size k for two phylogenetic trees is a forest which can be obtained from both trees by removing (k-1) edges in each tree and subsequently suppressing internal nodes of degree 2.
Acyclic agreement forests
A raffinement on the above definition can be made, resulting in the concept of acyclic agreement forest. An agreement forest F for two X-trees T1 and T2 is said to be acyclic if each of its tree components can be numbered in such a way that if the root of one component Xi ∈ F is an ancestor of the root of another component Xj ∈ F in either T1 or T2, then the number assigned to Xi is lower than the number assigned to Xj.
Another characterization of acyclicity in agreement forest is to consider the directed graph GF that has vertex set F and a directed edge (Xi, Xj) if and only if i ≠ j and at least one of the two following conditions hold:
- the root of T1(Xi) is an ancestor of the root of T1(Xj) in T1
- the root of T2(Xi) is an ancestor of the root of T2(Xj) in T2
The directed graph GF is called the inheritance graph associated with the agreement forest F, and we call F acyclic if GF has no directed cycle.

In computer science, an AVL tree is a self-balancing binary search tree. In an AVL tree, the heights of the two child subtrees of any node differ by at most one; if at any time they differ by more than one, rebalancing is done to restore this property. Lookup, insertion, and deletion all take O(log n) time in both the average and worst cases, where is the number of nodes in the tree prior to the operation. Insertions and deletions may require the tree to be rebalanced by one or more tree rotations.

In computer science, a binary tree is a k-ary tree data structure in which each node has at most two children, which are referred to as the left child and the right child. A recursive definition using just set theory notions is that a (non-empty) binary tree is a tuple (L, S, R), where L and R are binary trees or the empty set and S is a singleton set containing the root. Some authors allow the binary tree to be the empty set as well.

In computer science, a tree is a widely used abstract data type that represents a hierarchical tree structure with a set of connected nodes. Each node in the tree can be connected to many children, but must be connected to exactly one parent, except for the root node, which has no parent. These constraints mean there are no cycles or "loops", and also that each child can be treated like the root node of its own subtree, making recursion a useful technique for tree traversal. In contrast to linear data structures, many trees cannot be represented by relationships between neighboring nodes in a single straight line.

In graph theory, a tree is an undirected graph in which any two vertices are connected by exactly one path, or equivalently a connected acyclic undirected graph. A forest is an undirected graph in which any two vertices are connected by at most one path, or equivalently an acyclic undirected graph, or equivalently a disjoint union of trees.

A phylogenetic tree, phylogeny or evolutionary tree is a graphical representation which shows the evolutionary history between a set of species or taxa during a specific time. In another word, it is a branching diagram or a tree showing the evolutionary relationships among various biological species or other entities based upon similarities and differences in their physical or genetic characteristics. All life on Earth is part of a single phylogenetic tree, indicating common ancestry. Phylogenetics is the field of the study for the phylogenetic trees. The main challenge is to find a phylogenetic tree representing optimal evolutionary ancestry between a set of species or taxa. Computational phylogenetics focuses on the algorithms involved in finding optimal phylogenetic tree in the phylogenetic landscape.
In graph theory, a tree decomposition is a mapping of a graph into a tree that can be used to define the treewidth of the graph and speed up solving certain computational problems on the graph.

In computer science, the treap and the randomized binary search tree are two closely related forms of binary search tree data structures that maintain a dynamic set of ordered keys and allow binary searches among the keys. After any sequence of insertions and deletions of keys, the shape of the tree is a random variable with the same probability distribution as a random binary tree; in particular, with high probability its height is proportional to the logarithm of the number of keys, so that each search, insertion, or deletion operation takes logarithmic time to perform.
In computer science, a disjoint-set data structure, also called a union–find data structure or merge–find set, is a data structure that stores a collection of disjoint (non-overlapping) sets. Equivalently, it stores a partition of a set into disjoint subsets. It provides operations for adding new sets, merging sets, and finding a representative member of a set. The last operation makes it possible to find out efficiently if any two elements are in the same or different sets.
In computer science, a fusion tree is a type of tree data structure that implements an associative array on w-bit integers on a finite universe, where each of the input integers has size less than 2w and is non-negative. When operating on a collection of n key–value pairs, it uses O(n) space and performs searches in O(logwn) time, which is asymptotically faster than a traditional self-balancing binary search tree, and also better than the van Emde Boas tree for large values of w. It achieves this speed by using certain constant-time operations that can be done on a machine word. Fusion trees were invented in 1990 by Michael Fredman and Dan Willard.
Muller's method is a root-finding algorithm, a numerical method for solving equations of the form f(x) = 0. It was first presented by David E. Muller in 1956.
A skew heap is a heap data structure implemented as a binary tree. Skew heaps are advantageous because of their ability to merge more quickly than binary heaps. In contrast with binary heaps, there are no structural constraints, so there is no guarantee that the height of the tree is logarithmic. Only two conditions must be satisfied:
A propositional directed acyclic graph (PDAG) is a data structure that is used to represent a Boolean function. A Boolean function can be represented as a rooted, directed acyclic graph of the following form:
In constraint satisfaction, a decomposition method translates a constraint satisfaction problem into another constraint satisfaction problem that is binary and acyclic. Decomposition methods work by grouping variables into sets, and solving a subproblem for each set. These translations are done because solving binary acyclic problems is a tractable problem.
In computer science, weight-balanced binary trees (WBTs) are a type of self-balancing binary search trees that can be used to implement dynamic sets, dictionaries (maps) and sequences. These trees were introduced by Nievergelt and Reingold in the 1970s as trees of bounded balance, or BB[α] trees. Their more common name is due to Knuth.
Tree rearrangements are deterministic algorithms devoted to search for optimal phylogenetic tree structure. They can be applied to any set of data that are naturally arranged into a tree, but have most applications in computational phylogenetics, especially in maximum parsimony and maximum likelihood searches of phylogenetic trees, which seek to identify one among many possible trees that best explains the evolutionary history of a particular gene or species.
In mathematics, Newick tree format is a way of representing graph-theoretical trees with edge lengths using parentheses and commas. It was adopted by James Archie, William H. E. Day, Joseph Felsenstein, Wayne Maddison, Christopher Meacham, F. James Rohlf, and David Swofford, at two meetings in 1986, the second of which was at Newick's restaurant in Dover, New Hampshire, US. The adopted format is a generalization of the format developed by Meacham in 1984 for the first tree-drawing programs in Felsenstein's PHYLIP package.

Tarjan's strongly connected components algorithm is an algorithm in graph theory for finding the strongly connected components (SCCs) of a directed graph. It runs in linear time, matching the time bound for alternative methods including Kosaraju's algorithm and the path-based strong component algorithm. The algorithm is named for its inventor, Robert Tarjan.
In mathematics, specifically in graph theory and number theory, a hydra game is a single-player iterative mathematical game played on a mathematical tree called a hydra where, usually, the goal is to cut off the hydra's "heads" while the hydra simultaneously expands itself. Hydra games can be used to generate large numbers or infinite ordinals or prove the strength of certain mathematical theories.
In mathematics and computer science, an unrooted binary tree is an unrooted tree in which each vertex has either one or three neighbors.

The Euler tour technique (ETT), named after Leonhard Euler, is a method in graph theory for representing trees. The tree is viewed as a directed graph that contains two directed edges for each edge in the tree. The tree can then be represented as a Eulerian circuit of the directed graph, known as the Euler tour representation (ETR) of the tree. The ETT allows for efficient, parallel computation of solutions to common problems in algorithmic graph theory. It was introduced by Tarjan and Vishkin in 1984.









