Related Research Articles
Molecular biology is a branch of biology that seeks to understand the molecular basis of biological activity in and between cells, including biomolecular synthesis, modification, mechanisms, and interactions.

The polymerase chain reaction (PCR) is a method widely used to make millions to billions of copies of a specific DNA sample rapidly, allowing scientists to amplify a very small sample of DNA sufficiently to enable detailed study. PCR was invented in 1983 by American biochemist Kary Mullis at Cetus Corporation. Mullis and biochemist Michael Smith, who had developed other essential ways of manipulating DNA, were jointly awarded the Nobel Prize in Chemistry in 1993.
Viral load, also known as viral burden, is a numerical expression of the quantity of virus in a given volume of fluid, including biological and environmental specimens. It is not to be confused with viral titre or viral titer, which depends on the assay. When an assay for measuring the infective virus particle is done, viral titre often refers to the concentration of infectious viral particles, which is different from the total viral particles. Viral load is measured using body fluids sputum and blood plasma. As an example of environmental specimens, the viral load of norovirus can be determined from run-off water on garden produce. Norovirus has not only prolonged viral shedding and has the ability to survive in the environment but a minuscule infectious dose is required to produce infection in humans: less than 100 viral particles.

DNA computing is an emerging branch of unconventional computing which uses DNA, biochemistry, and molecular biology hardware, instead of the traditional electronic computing. Research and development in this area concerns theory, experiments, and applications of DNA computing. Although the field originally started with the demonstration of a computing application by Len Adleman in 1994, it has now been expanded to several other avenues such as the development of storage technologies, nanoscale imaging modalities, synthetic controllers and reaction networks, etc.

Ribonuclease H is a family of non-sequence-specific endonuclease enzymes that catalyze the cleavage of RNA in an RNA/DNA substrate via a hydrolytic mechanism. Members of the RNase H family can be found in nearly all organisms, from bacteria to archaea to eukaryotes.
Deoxyribozymes, also called DNA enzymes, DNAzymes, or catalytic DNA, are DNA oligonucleotides that are capable of performing a specific chemical reaction, often but not always catalytic. This is similar to the action of other biological enzymes, such as proteins or ribozymes . However, in contrast to the abundance of protein enzymes in biological systems and the discovery of biological ribozymes in the 1980s, there is only little evidence for naturally occurring deoxyribozymes. Deoxyribozymes should not be confused with DNA aptamers which are oligonucleotides that selectively bind a target ligand, but do not catalyze a subsequent chemical reaction.

QIAGEN N.V. is a German-founded multinational provider of sample and assay technologies for molecular diagnostics, applied testing, academic research, and pharmaceutical research. The company operates in more than 35 offices in over 25 countries. QIAGEN N.V., the global corporate headquarter of the QIAGEN group, is located in Venlo, The Netherlands. The main operative headquarters are located in Hilden, Germany. European, American, Chinese, and Asian-Pacific regional headquarters are located respectively in respectively Hilden, Germany; Germantown, Maryland, United States; Shanghai, China; and Singapore. QIAGEN's shares are listed at the NYSE and at the Frankfurt Stock Exchange in the Prime Standard. Thierry Bernard is the company's Chief Executive Officer (CEO).
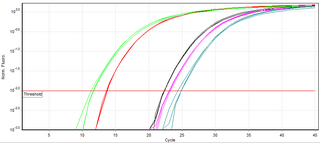
A real-time polymerase chain reaction is a laboratory technique of molecular biology based on the polymerase chain reaction (PCR). It monitors the amplification of a targeted DNA molecule during the PCR, not at its end, as in conventional PCR. Real-time PCR can be used quantitatively and semi-quantitatively.
Dz13 is an experimental treatment developed by scientists at the University of New South Wales. The drug aims to combat a range of illnesses, including skin cancer, restenosis, arthritis and macular degeneration. Trials of Dz13 were suspended in 2013.
In biology, a branched DNA assay is a signal amplification assay that is used to detect nucleic acid molecules.

Systematic evolution of ligands by exponential enrichment (SELEX), also referred to as in vitro selection or in vitro evolution, is a combinatorial chemistry technique in molecular biology for producing oligonucleotides of either single-stranded DNA or RNA that specifically bind to a target ligand or ligands. These single-stranded DNA or RNA are commonly referred to as aptamers. Although SELEX has emerged as the most commonly used name for the procedure, some researchers have referred to it as SAAB and CASTing SELEX was first introduced in 1990. In 2015, a special issue was published in the Journal of Molecular Evolution in the honor of quarter century of the discovery of SELEX.
Nucleic acid sequence-based amplification, commonly referred to as NASBA, is a method in molecular biology which is used to produce multiple copies of single stranded RNA. NASBA is a two-step process that takes RNA and anneals specially designed primers, then utilizes an enzyme cocktail to amplify it.
Digital polymerase chain reaction is a biotechnological refinement of conventional polymerase chain reaction methods that can be used to directly quantify and clonally amplify nucleic acids strands including DNA, cDNA, or RNA. The key difference between dPCR and qPCR lies in the method of measuring nucleic acids amounts, with the former being a more precise method than PCR, though also more prone to error in the hands of inexperienced users. PCR carries out one reaction per single sample. dPCR also carries out a single reaction within a sample, however the sample is separated into a large number of partitions and the reaction is carried out in each partition individually. This separation allows a more reliable collection and sensitive measurement of nucleic acid amounts. The method has been demonstrated as useful for studying variations in gene sequences—such as copy number variants and point mutations.
Biomolecular engineering is the application of engineering principles and practices to the purposeful manipulation of molecules of biological origin. Biomolecular engineers integrate knowledge of biological processes with the core knowledge of chemical engineering in order to focus on molecular level solutions to issues and problems in the life sciences related to the environment, agriculture, energy, industry, food production, biotechnology and medicine.
Integrated DNA Technologies, Inc. (IDT), headquartered in Coralville, Iowa, is a supplier of custom nucleic acids, serving the areas of academic research, biotechnology, clinical diagnostics, and pharmaceutical development. IDT's primary business is the manufacturing of custom DNA and RNA oligonucleotides (oligos) for research applications.
Hot start PCR is a modified form of conventional polymerase chain reaction (PCR) that reduces the presence of undesired products and primer dimers due to non-specific DNA amplification at room temperatures. Many variations and modifications of the PCR procedure have been developed in order to achieve higher yields; hot start PCR is one of them. Hot start PCR follows the same principles as the conventional PCR - in that it uses DNA polymerase to synthesise DNA from a single stranded template. However, it utilizes additional heating and separation methods, such as inactivating or inhibiting the binding of Taq polymerase and late addition of Taq polymerase, to increase product yield as well as provide a higher specificity and sensitivity. Non-specific binding and priming or formation of primer dimers are minimized by completing the reaction mix after denaturation. Some ways to complete reaction mixes at high temperatures involve modifications that block DNA polymerase activity in low temperatures, use of modified deoxyribonucleotide triphosphates (dNTPs), and the physical addition of one of the essential reagents after denaturation.
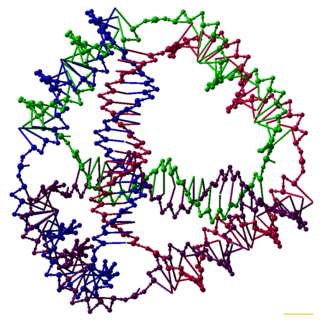
DNA nanotechnology is the design and manufacture of artificial nucleic acid structures for technological uses. In this field, nucleic acids are used as non-biological engineering materials for nanotechnology rather than as the carriers of genetic information in living cells. Researchers in the field have created static structures such as two- and three-dimensional crystal lattices, nanotubes, polyhedra, and arbitrary shapes, and functional devices such as molecular machines and DNA computers. The field is beginning to be used as a tool to solve basic science problems in structural biology and biophysics, including applications in X-ray crystallography and nuclear magnetic resonance spectroscopy of proteins to determine structures. Potential applications in molecular scale electronics and nanomedicine are also being investigated.
Recombinase polymerase amplification (RPA) is a single tube, isothermal alternative to the polymerase chain reaction (PCR). By adding a reverse transcriptase enzyme to an RPA reaction, it can detect RNA as well as DNA, without the need for a separate step to produce cDNA. Because it is isothermal, RPA can use much simpler equipment than PCR, which requires a thermal cycler. Operating best at temperatures of 37–42 °C and still working, albeit more slowly, at room temperature means RPA reactions can in theory be run quickly by simply holding a tube in the hand. This makes RPA an excellent candidate for developing low-cost, rapid, point-of-care molecular tests. An international quality assessment of molecular detection of Rift Valley fever virus performed as well as the best RT-PCR tests, detecting less concentrated samples missed by some PCR tests and an RT-LAMP test. RPA was developed and launched by TwistDx Ltd., a biotechnology company based in Cambridge, UK.

Molecular diagnostics is a collection of techniques used to analyze biological markers in the genome and proteome, and how their cells express their genes as proteins, applying molecular biology to medical testing. In medicine the technique is used to diagnose and monitor disease, detect risk, and decide which therapies will work best for individual patients, and in agricultural biosecurity similarly to monitor crop- and livestock disease, estimate risk, and decide what quarantine measures must be taken.
Diagnostic microbiology is the study of microbial identification. Since the discovery of the germ theory of disease, scientists have been finding ways to harvest specific organisms. Using methods such as differential media or genome sequencing, physicians and scientists can observe novel functions in organisms for more effective and accurate diagnosis of organisms. Methods used in diagnostic microbiology are often used to take advantage of a particular difference in organisms and attain information about what species it can be identified as, which is often through a reference of previous studies. New studies provide information that others can reference so that scientists can attain a basic understanding of the organism they are examining.
References
- ↑ Todd, Alison Velyian (1992), Molecular analysis of regulatory and transforming sequences of the human N-ras gene
- ↑ "News - Brisbane Girls Grammar School". www.bggs.qld.edu.au. Retrieved 2 August 2019.
- 1 2 "SpeeDx | Executive Team". SpeeDx. Archived from the original on 19 July 2019. Retrieved 27 July 2019.
- ↑ "True Leaders Game Changers 2017: SpeeDx's Alison Todd frontline fighter in the war on superbugs". Australian Financial Review. 12 October 2017. Retrieved 27 July 2019.
- ↑ Commission, Australian Trade and Investment. "Australian startup wages war on drug-resistant sup". www.austrade.gov.au. Retrieved 27 July 2019.
- 1 2 "Dr Alison Todd returns to Girls Grammar to share her passion for entrepreneurship - Brisbane Girls Grammar School". www.bggs.qld.edu.au. Retrieved 2 August 2019.
- ↑ "Australian Biochemist" (PDF). 25 July 2019. Archived from the original (PDF) on 20 October 2022.
- ↑ Santoro, S. W.; Joyce, G. F. (29 April 1997). "A general purpose RNA-cleaving DNA enzyme". Proceedings of the National Academy of Sciences of the United States of America. 94 (9): 4262–4266. Bibcode:1997PNAS...94.4262S. doi: 10.1073/pnas.94.9.4262 . ISSN 0027-8424. PMC 20710 . PMID 9113977.
- ↑ Todd, A. V.; Fuery, C. J.; Impey, H. L.; Applegate, T. L.; Haughton, M. A. (May 2000). "DzyNA-PCR: use of DNAzymes to detect and quantify nucleic acid sequences in a real-time fluorescent format". Clinical Chemistry. 46 (5): 625–630. doi: 10.1093/clinchem/46.5.625 . ISSN 0009-9147. PMID 10794743.
- ↑ Mokany, Elisa; Bone, Simon M.; Young, Paul E.; Doan, Tram B.; Todd, Alison V. (27 January 2010). "MNAzymes, a Versatile New Class of Nucleic Acid Enzymes That Can Function as Biosensors and Molecular Switches". Journal of the American Chemical Society. 132 (3): 1051–1059. doi:10.1021/ja9076777. ISSN 0002-7863. PMC 2808728 . PMID 20038095.
- ↑ Todd, Alison V.; Fuery, Caroline J.; Bone, Simon M.; Tan, Yee Lee; Mokany, Elisa (1 February 2013). "MNAzyme qPCR with Superior Multiplexing Capacity". Clinical Chemistry. 59 (2): 419–426. doi: 10.1373/clinchem.2012.192930 . ISSN 0009-9147. PMID 23232065.
- ↑ "Alison Todd — Google Scholar Citations". scholar.google.com. Retrieved 27 July 2019.
- ↑ "SpeeDx | Executive Team". SpeeDx. Archived from the original on 19 July 2019. Retrieved 28 July 2019.
- ↑ "Dr Alison Todd – Molecular inventor". Applied. Retrieved 24 October 2019.
- ↑ "True Leaders Game Changers 2017: SpeeDx's Alison Todd frontline fighter in the war on superbugs". Australian Financial Review. 12 October 2017. Retrieved 27 July 2019.
- ↑ Khadem, Nassim (27 November 2017). "Aussie company SpeeDx is fighting a sexually transmitted infection that's acting like a superbug". The Sydney Morning Herald. Retrieved 27 July 2019.