
Ribosomes comprise a complex macromolecular machine, found within all living cells, that serves as the site of biological protein synthesis (translation). Ribosomes link amino acids together in the order specified by messenger RNA (mRNA) molecules. Ribosomes consist of two major components: the small ribosomal subunits, which read the mRNA, and the large subunits, which join amino acids to form a polypeptide chain. Each subunit consists of one or more ribosomal RNA (rRNA) molecules and a variety of ribosomal proteins. The ribosomes and associated molecules are also known as the translational apparatus.
The 5′ untranslated region is the region of an mRNA that is directly upstream from the initiation codon. This region is important for the regulation of translation of a transcript by differing mechanisms in viruses, prokaryotes and eukaryotes. While called untranslated, the 5′ UTR or a portion of it is sometimes translated into a protein product. This product can then regulate the translation of the main coding sequence of the mRNA. In many organisms, however, the 5′ UTR is completely untranslated, instead forming complex secondary structure to regulate translation. The 5′ UTR has been found to interact with proteins relating to metabolism; and proteins translate sequences within the 5′ UTR. In addition, this region has been involved in transcription regulation, such as the sex-lethal gene in Drosophila. Regulatory elements within 5′ UTRs have also been linked to mRNA export.
Prokaryotic translation is the process by which messenger RNA is translated into proteins in prokaryotes.
Eukaryotic translation is the biological process by which messenger RNA is translated into proteins in eukaryotes. It consists of four phases: initiation, elongation, termination, and recycling.
The Kozak consensus sequence, Kozak consensus or Kozak sequence is a sequence which occurs on eukaryotic mRNA and has the consensus (gcc)gccRccAUGG. The Kozak consensus sequence plays a major role in the initiation of the translation process. The sequence was named after the scientist who discovered it, Marilyn Kozak.

The BiP internal ribosome entry site (IRES) is an RNA element present in the 5' UTR of the mRNA of BiP protein and allows cap-independent translation. BiP protein expression has been found to be significantly enhanced by the heat shock response due to internal ribosome entry site (IRES)-dependent translation. It is thought that this translational mechanism is essential for the survival of cells under stress.

The Cripavirus internal ribosome entry site is an RNA element required for the production of capsid proteins through ribosome recruitment to an intergenic region IRES.

This family represents the internal ribosome entry site (IRES) of the hepatitis A virus. HAV IRES is a 450 nucleotide long sequence located in the 735 nt long 5’ UTR of Hepatitis A viral RNA genome. IRES elements allow cap and end-independent translation of mRNA in the host cell. The IRES achieves this by mediating the internal initiation of translation by recruiting a ribosomal 40S pre-initiation complex directly to the initiation codon and eliminates the requirement for eukaryotic initiation factor, eIF4F.
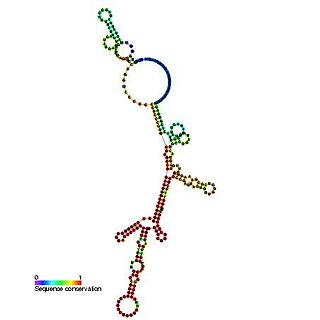
The Hepatitis C virus internal ribosome entry site, or HCV IRES, is an RNA structure within the 5'UTR of the HCV genome that mediates cap-independent translation initiation.
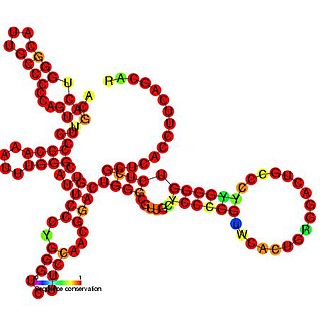
The insulin-like growth factor II (IGF-II) internal ribosome entry site IRES is found in the 5' UTR of IGF-II leader 2 mRNA. This RNA element allows cap-independent translation of the mRNA and it is thought that this family may facilitate a continuous IGF-II production in rapidly dividing cells during development. Ribosomal scanning on human insulin-like growth factor II (IGF-II) is hard to comprehend due to one open reading frame and the ability for the hormone to fold into a stable structure.

This family represents the internal ribosome entry site (IRES) of the pestiviruses. The pestivirus IRES allows cap and end-independent translation of mRNA in the host cell. The IRES achieves this by mediating the internal initiation of translation by recruiting a ribosomal 43S pre-initiation complex directly to the initiation codon and eliminates the requirement for the eukaryotic initiation factor, eIF4F. The classical swine fever virus UTR described appears to be longer at the 5' end than other pestivirus UTRs. This family represents the conserved core.
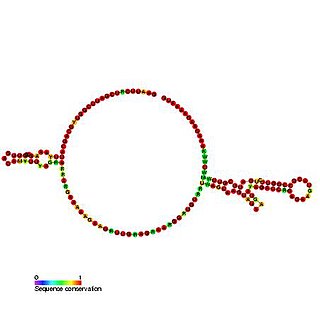
The Tobamovirus internal ribosome entry site (IRES) is an element that allows cap and end-independent translation of mRNA in the host cell. The IRES achieves this by mediating the internal initiation of translation by recruiting a ribosomal 43S pre-initiation complex directly to the initiation codon and eliminates the requirement for the eukaryotic initiation factor, eIF4F.

The TrkB internal ribosome entry site (IRES) is an RNA element which is present in the 5' UTR sequence of the mRNA. TrkB is a neurotrophin receptor which is essential for the development and maintenance of the nervous system. The internal ribosome entry site IRES element allows cap-independent translation of TrkB which may be needed for efficient translation in neuronal dendrites.
A ribosome binding site, or ribosomal binding site (RBS), is a sequence of nucleotides upstream of the start codon of an mRNA transcript that is responsible for the recruitment of a ribosome during the initiation of protein translation. Mostly, RBS refers to bacterial sequences, although internal ribosome entry sites (IRES) have been described in mRNAs of eukaryotic cells or viruses that infect eukaryotes. Ribosome recruitment in eukaryotes is generally mediated by the 5' cap present on eukaryotic mRNAs.
The eukaryotic small ribosomal subunit (40S) is the smaller subunit of the eukaryotic 80S ribosomes, with the other major component being the large ribosomal subunit (60S). The "40S" and "60S" names originate from the convention that ribosomal particles are denoted according to their sedimentation coefficients in Svedberg units. It is structurally and functionally related to the 30S subunit of 70S prokaryotic ribosomes. However, the 40S subunit is much larger than the prokaryotic 30S subunit and contains many additional protein segments, as well as rRNA expansion segments.
Eukaryotic translation initiation factor 4 G (eIF4G) is a protein involved in eukaryotic translation initiation and is a component of the eIF4F cap-binding complex. Orthologs of eIF4G have been studied in multiple species, including humans, yeast, and wheat. However, eIF4G is exclusively found in domain Eukarya, and not in domains Bacteria or Archaea, which do not have capped mRNA. As such, eIF4G structure and function may vary between species, although the human eIF4G 1 has been the focus of extensive studies.
The P-site is the second binding site for tRNA in the ribosome. The other two sites are the A-site (aminoacyl), which is the first binding site in the ribosome, and the E-site (exit), is the third and final binding site in the ribosome. During protein translation, the P-site holds the tRNA which is linked to the growing polypeptide chain. When a stop codon is reached, the peptidyl-tRNA bond of the tRNA located in the P-site is cleaved releasing the newly synthesized protein. During the translocation step of the elongation phase, the mRNA is advanced by one codon, coupled to movement of the tRNAs from the ribosomal A to P and P to E sites, catalyzed by elongation factor EF-G.
In molecular biology, the Ure2 internal ribosome entry site (IRES) is an RNA element present in the 5' UTR of the mRNA of Ure2. It allows 5' cap- and eIF4E-independent translation of an N-terminally truncated form of Ure2. This truncated form lacks the prion-forming domain. It is a 104 nucleotide region, smaller than most viral IRES elements, which forms a stem-loop structure. EIF2A represses this IRES resulting in an inhibition of translation of the N-terminally truncated Ure2.










