
The SAM-II riboswitch is an RNA element found predominantly in Alphaproteobacteria that binds S-adenosyl methionine (SAM). Its structure and sequence appear to be unrelated to the SAM riboswitch found in Gram-positive bacteria. This SAM riboswitch is located upstream of the metA and metC genes in Agrobacterium tumefaciens, and other methionine and SAM biosynthesis genes in other alpha-proteobacteria. Like the other SAM riboswitch, it probably functions to turn off expression of these genes in response to elevated SAM levels. A significant variant of SAM-II riboswitches was found in Pelagibacter ubique and related marine bacteria and called SAM-V. Also, like many structured RNAs, SAM-II riboswitches can tolerate long loops between their stems.

The Acido-1 RNA motif is a conserved RNA structure identified by bioinformatics. It is found only in acidobacteriota, and appears to be a non-coding RNA as it does not have a consistent association with protein-coding genes.

The Actino-pnp RNA motif is a conserved structure found in Actinomycetota that is apparently in the 5' untranslated regions of genes predicted to encode exoribonucleases. The RNA element's function is likely analogous to an RNA structure found upstream of polynucleotide phosphorylase genes in E. coli and related enterobacteria. In this latter system, the polynucleotide phosphorlyase gene regulates its own expression levels by a feedback mechanism that involves its activity upon the RNA structure. However, the E. coli RNA appears to be structurally unrelated to the Actino-pnp motif.

The Bacillaceae-1 RNA motif is a conserved RNA structure identified by bioinformatics within bacteria in the family bacillaceae. The RNA is presumed to operate as a non-coding RNA, and is sometimes adjacent to operons containing ribosomal RNAs. The most characteristic feature is two terminal loops that have the nucleotide consensus RUCCU, where R is either A or G. The motif might be related to the Desulfotalea-1 RNA motif, as the motifs share some similarity in conserved features, and the Desulfotalea-1 RNA motif is also sometimes adjacent to ribosomal RNA operons.

The Bacteroides-1 RNA motif is a conserved RNA structure identified in bacteria within the genus Bacteroides. The RNAs are typically found downstream of of genes that participate in the synthesis of exopolysaccharides of unknown types. It is possible that Bacteroides-1 RNAs regulate the upstream genes, but since this mode of regulation is unusual in bacteria, it is more likely that the structure functions as a non-coding RNA.

The Chloroflexi-1 RNA motif is a conserved RNA structure detected by bioinformatics within the species Chloroflexus aggregans. C. aggregans has three predicted Chloroflexi-1 RNAs, which are located nearby to one another. This arrangement might suggest a repetitive element. C. aggregans is classified as belonging to the bacterial phylum Chloroflexota.

The Collinsella-1 RNA motif denotes a particular conserved RNA structure discovered by bioinformatics. Of the six sequences belonging to this motif that were originally identified, five are from uncultivated bacteria residing in the human gut, while only the sixth is in a cultivated species, Collinsella aerofaciens. The evidence supporting the stem-loops designated as "P1" and "P2" is ambiguous.
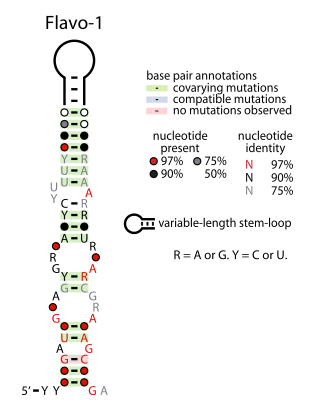
The Flavo-1 RNA motif is a conserved RNA structure that was identified by bioinformatics. The vast majority of Flavo-1 RNAs are found in Flavobacteria, but some were detected in the phylum Bacteroidota, which contains Flavobacteria, or the phylum Spirochaetota, which is evolutionarily related to Bacteroidota. It was presumed that Flavo-1 RNAs function as non-coding RNAs.
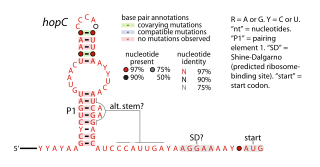
The hopC RNA motif is a predicted cis-regulatory element identified by a bioinformatic screen for conserved RNA secondary structures. hopC RNAs are exclusively found within bacteria classified within the genus Helicobacter, some of which are human pathogens that infect the stomach and can cause ulcers.
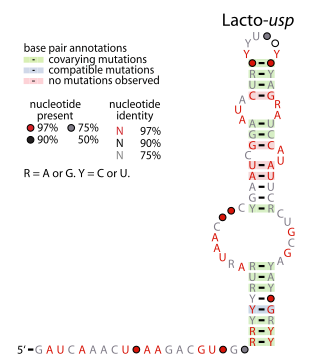
The Lacto-usp RNA motif is a conserved RNA structure identified in bacteria by bioinformatics. Lacto-usp RNAs are found exclusively in lactic acid bacteria, and exclusively in the possible 5′ untranslated regions of operons that contain a hypothetical gene and a usp gene. The usp gene encodes the universal stress protein. It was proposed that the Lacto-usp might correspond to the 6S RNA of the relevant species, because four of five of these species lack a predicted 6S RNA, and 6S RNAs commonly occur in 5′ UTRs of usp genes. However, given that the Lacto-usp RNA motif is much shorter than the standard 6S RNA structure, the function of Lacto-usp RNAs remains unclear.

The Moco-II RNA motif is a conserved RNA structure identified by bioinformatics. However, only 8 examples of the RNA motif are known. The RNAs are potentially in the 5' untranslated regions of genes related to molybdenum cofactor (Moco), specifically a gene that encodes a molybdenum-binding domain and a nitrate reductase, which uses Moco as a cofactor. Thus the RNA might be involved in the regulation of genes based on Moco levels. Reliable predictions of Moco-II RNAs are restricted to deltaproteobacteria, but a Moco-II RNA might be present in a betaproteobacterial species. The Moco RNA motif is another RNA that is associated with Moco, and its complex secondary structure and genetic experiments have led to proposals that it is a riboswitch. However, the simpler structure of the Moco-II RNA motif is less typical of riboswitches. Moco-II RNAs are typically followed by a predicted rho-independent transcription terminator.
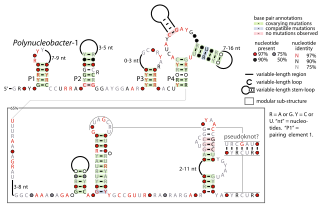
The Polynucleobacter-1 RNA motif is a conserved RNA structure that was identified by bioinformatics. The RNA structure is predominantly located in genome sequences derived from DNA extracted from uncultivated marine samples. However it was also predicted in the genome of Polynucleobacter species QLW-P1DMWA-1, a kind of betaproteobacteria. The RNAs are often located near to a conserved gene that might be homologous to a gene found in a phage that infects cyanobacteria. However, it is unknown if the RNA is used by phages.
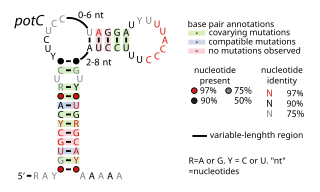
The potC RNA motif is a conserved RNA structure discovered using bioinformatics. The RNA is detected only in genome sequences derived from DNA that was extracted from uncultivated marine bacteria. Thus, this RNA is present in environmental samples, but not yet found in any cultivated organism. potC RNAs are located in the presumed 5' untranslated regions of genes predicted to encode either membrane transport proteins or peroxiredoxins. Therefore, it was hypothesized that potC RNAs are cis-regulatory elements, but their detailed function is unknown.
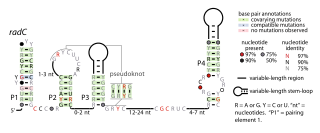
The radC RNA motif is a conserved RNA structure identified by bioinformatics. The radC RNA motif is found in certain bacteria where it is consistent located in the presumed 5' untranslated regions of genes whose encoded proteins bind DNA are interact with other proteins that bind DNA. These proteins include integrases, methyltransferases that might methylate DNA, proteins that inhibit restriction enzymes and radC genes. Although radC genes were thought to encode DNA repair proteins, this conclusion was based on mutation data that was later shown to affect a different gene. However, it is still possible that radC genes play some DNA-related role. No radC RNAs have been detected in any purified phage whose sequence was available as of 2010, although integrases are often used by phages.

The Ssbp, Topoisomerase, Antirestriction, XerDC Integrase RNA motif is a conserved RNA-like structure identified using bioinformatics. STAXI RNAs are located near to genes encoding proteins that interact with DNA or are associated with such proteins. This observation raised the possibility that instances of the STAXI motif function as single-stranded DNA molecules, perhaps during DNA replication or DNA repair. On the other hand, STAXI motifs often contain terminal loops conforming to the stable UNCG tetraloop, but the DNA version of this tetraloop (TNCG) is not especially stable. The STAXI motif consists of a simple pseudoknot structure that is repeated two or more times.
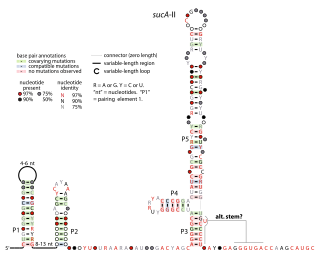
The sucA-II RNA motif is a conserved RNA structure identified by bioinformatics. It is consistently found in the presumed 5' untranslated regions of sucA genes, which encode Oxoglutarate dehydrogenase enzymes that participate in the citric acid cycle. Given this arrangement, sucA-II RNAs might regulate the downstream sucA gene. This genetic arrangement is similar to the previously reported sucA RNA motif. However, sucA-II RNAs are found only in bacteria classified within the genus Pseudomonas, whereas the previously reported motif is found only in betaproteobacteria.

The sucC RNA motif is a conserved RNA structure discovered using bioinformatics. sucC RNAs are found in the genus Pseudomonas, and are consistently found in possible 5' untranslated regions of sucC genes. These genes encode Succinyl coenzyme A synthetase, and are hypothesised to be regulated by the sucC RNAs. sucC genes participate in the citric acid cycle, and another gene involved in the citric acid cycle, sucA, is also predicted to be regulated by a conserved RNA structure.
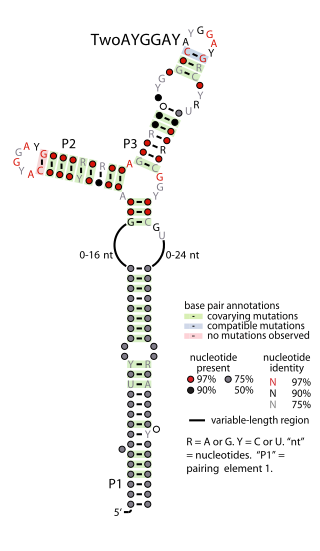
The TwoAYGGAY RNA motif is a conserved RNA structure identified by bioinformatics. Its name refers to the conserved AYGGAY nucleotide sequence found in the motif's two terminal loops. The RNAs are found in sequences derived from DNA extracted from uncultivated bacteria present in the human gut, as well as some bacteria in the classes Clostridia and Gammaproteobacteria.
The Ocean-V RNA motif is a conserved RNA structure discovered using bioinformatics. Only a few Ocean-V RNA sequences have been detected, all in sequences derived from DNA that was extracted from uncultivated bacteria found in ocean water. As of 2010, no Ocean-V RNA has been detected in any known, cultivated organism.

The eps-Associated RNA element is a conserved RNA motif associated with exopolysaccharide (eps) or capsule biosynthesis genes in a subset of bacteria classified within the order Bacillales. It was initially discovered in Bacillus subtilis, located between the second and third gene in the eps operon. Deletion of the EAR element impairs biofilm formation.



















