
A protein kinase is a kinase enzyme that modifies other molecules, mostly proteins, by chemically adding phosphate groups to them (phosphorylation). Phosphorylation usually results in a functional change of the target protein (substrate) by changing enzyme activity, cellular location, or association with other proteins. The human genome contains about 518 protein kinase genes and they constitute about 2% of all human genes. Up to 30% of all human proteins may be modified by kinase activity, and kinases are known to regulate the majority of cellular pathways, especially those involved in signal transduction. Protein kinases are also found in bacteria and plants, and include the pseudokinase sub-family, which exhibit unusual features including atypical nucleotide binding and weak, or no, catalytic activity and are part of a much larger pseudoenzyme group of 'degraded' enzyme relatives that are found throughout life, where they take an active participation in mechanistic cellular signaling.
Bart is a masculine given name and a surname.

In the field of molecular biology, a two-component regulatory system serves as a basic stimulus-response coupling mechanism to allow organisms to sense and respond to changes in many different environmental conditions. Two-component systems typically consist of a membrane-bound histidine kinase that senses a specific environmental stimulus and a corresponding response regulator that mediates the cellular response, mostly through differential expression of target genes. Although two-component signaling systems are found in all domains of life, they are most common by far in bacteria, particularly in Gram-negative and cyanobacteria; both histidine kinases and response regulators are among the largest gene families in bacteria. They are much less common in archaea and eukaryotes; although they do appear in yeasts, filamentous fungi, and slime molds, and are common in plants, two-component systems have been described as "conspicuously absent" from animals.
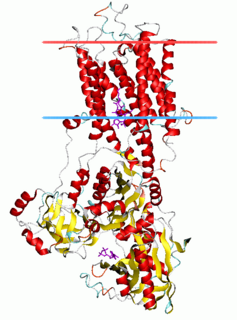
The P-type ATPases, also known as E1-E2 ATPases, are a large group of evolutionarily related ion and lipid pumps that are found in bacteria, archaea, and eukaryotes. P-type ATPases are α-helical bundle primary transporters named based upon their ability to catalyze auto- (or self-) phosphorylation (hence P) of a key conserved aspartate residue within the pump and their energy source, adenosine triphosphate (ATP). In addition, they all appear to interconvert between at least two different conformations, denoted by E1 and E2. P-type ATPases fall under the P-type ATPase (P-ATPase) Superfamily (TC# 3.A.3) which, as of early 2016, includes 20 different protein families.

Histidine kinases (HK) are multifunctional, and in non-animal kingdoms, typically transmembrane, proteins of the transferase class of enzymes that play a role in signal transduction across the cellular membrane. The vast majority of HKs are homodimers that exhibit autokinase, phosphotransfer, and phosphatase activity. HKs can act as cellular receptors for signaling molecules in a way analogous to tyrosine kinase receptors (RTK). Multifunctional receptor molecules such as HKs and RTKs typically have portions on the outside of the cell that bind to hormone- or growth factor-like molecules, portions that span the cell membrane, and portions within the cell that contain the enzymatic activity. In addition to kinase activity, the intracellular domains typically have regions that bind to a secondary effector molecule or complex of molecules that further propagate signal transduction within the cell. Distinct from other classes of protein kinases, HKs are usually parts of a two-component signal transduction mechanisms in which HK transfers a phosphate group from ATP to a histidine residue within the kinase, and then to an aspartate residue on the receiver domain of a response regulator protein. More recently, the widespread existence of protein histidine phosphorylation distinct from that of two-component histidine kinases has been recognised in human cells. In marked contrast to Ser, Thr and Tyr phosphorylation, the analysis of phosphorylated Histidine using standard biochemical and mass spectrometric approaches is much more challenging, and special procedures and separation techniques are required for their preservation alongside classical Ser, Thr and Tyr phosphorylation on proteins isolated from human cells.
In enzymology, a polyphosphate kinase, or polyphosphate polymerase, is an enzyme that catalyzes the formation of polyphosphate from ATP, with chain lengths of up to a thousand or more orthophosphate moieties.
The Nucleobase:Cation Symporter-1 (NCS1) Family (TC# 2.A.39) consists of over 1000 currently sequenced proteins derived from Gram-negative and Gram-positive bacteria, archaea, fungi and plants. These proteins function as transporters for nucleobases including purines and pyrimidines. Members of this family possess twelve transmembrane α-helical spanners (TMSs). At least some of them have been shown to function in uptake by substrate:H+ symport mechanism.

Members of the Solute:Sodium Symporter (SSS) Family (TC# 2.A.21) catalyze solute:Na+ symport. The SSS family is within the APC Superfamily. The solutes transported may be sugars, amino acids, organo cations such as choline, nucleosides, inositols, vitamins, urea or anions, depending on the system. Members of the SSS family have been identified in bacteria, archaea and eukaryotes. Almost all functionally well-characterized members normally catalyze solute uptake via Na+ symport.
In molecular biology, the HAMP domain is an approximately 50-amino acid alpha-helical region that forms a dimeric, four-helical coiled coil. It is found in bacterial sensor and chemotaxis proteins and in eukaryotic histidine kinases. The bacterial proteins are usually integral membrane proteins and part of a two-component signal transduction pathway. One or several copies of the HAMP domain can be found in association with other domains, such as the histidine kinase domain, the bacterial chemotaxis sensory transducer domain, the PAS repeat, the EAL domain, the GGDEF domain, the protein phosphatase 2C-like domain, the guanylate cyclase domain, or the response regulatory domain. In its most common setting, the HAMP domain transmits conformational changes in periplasmic ligand-binding domains to cytoplasmic signalling kinase and methyl-acceptor domains and thus regulates the phosphorylation or methylation activity of homodimeric receptors.
The amino acid-polyamine-organocation (APC) superfamily is the second largest superfamily of secondary carrier proteins currently known, and it contain several Solute carriers. Originally, the APC superfamily consisted of subfamilies under the transporter classification number 2.A.3. This superfamily has since been expanded to include eighteen different families.
Lysine Exporters are a superfamily of transmembrane proteins which export amino acids, lipids and heavy metal ions. They provide ionic homeostasis, play a role in cell envelope assembly, and protect from excessive concentrations of heavy metals in cytoplasm. The superfamily was named based on the early discovery of the LysE carrier protein of Corynebacterium glutamicum.
Arsenite-antimonite transporters are membrane transporters that pump arsenite or antimonite out of a cell. Antimonite is the salt of antimony and has been found to significantly impact the toxicity of arsenite. The similar structure of As(III) and Sb(III) makes it plausible that certain transporters function in the efflux of both substrates. Arsenic efflux transporters exist in almost every organism and serve to remove this toxic compound from the cell.
Divalent anion:Na+ symporters were found in bacteria, archaea, plant chloroplasts and animals.
Arsenite resistance (Ars) efflux pumps of bacteria may consist of two proteins, ArsB and ArsA, or of one protein. ArsA proteins have two ATP binding domains and probably arose by a tandem gene duplication event. ArsB proteins all possess twelve transmembrane spanners and may also have arisen by a tandem gene duplication event. Structurally, the Ars pumps resemble ABC-type efflux pumps, but there is no significant sequence similarity between the Ars and ABC pumps. When only ArsB is present, the system operates by a pmf-dependent mechanism, and consequently belongs in TC subclass 2.A. When ArsA is also present, ATP hydrolysis drives efflux, and consequently the system belongs in TC subclass 3.A. ArsB therefore appears twice in the TC system but ArsA appears only once. These pumps actively expel both arsenite and antimonite.
The arsenical resistance-3 (ACR3) family is a member of the BART superfamily. Based on operon analyses, ARC3 homologues may function either as secondary carriers or as primary active transporters, similarly to the ArsB and ArsAB families. In the latter case ATP hydrolysis again energizes transport. ARC3 homologues transport the same anions as ArsA/AB homologues, though ArsB homologues are members of the IT Superfamily and homologues of the ARC3 family are within the BART Superfamily suggesting they may not be evolutionarily related.
The Holin superfamily IV is a superfamily of integral membrane transport proteins. It is one of the seven different holin superfamilies in total.
The Holin superfamily VI is a superfamily of integral membrane transport proteins. It is one of the seven different holin superfamilies in total. In general, these proteins are thought to play a role in regulated cell death, although functionality varies between families and individual members.
The K+Transporter (Trk) Family is a member of the voltage-gated ion channel (VIC) superfamily. The proteins of the Trk family are derived from Gram-negative and Gram-positive bacteria, yeast and plants.
YedZ of E. coli has been examined topologically and has 6 transmembrane segments (TMSs) with both the N- and C-termini localized to the cytoplasm. von Rozycki et al. 2004 identified homologues of YedZ in bacteria and animals. YedZ homologues exhibit conserved histidyl residues in their transmembrane domains that may function in heme binding. Some of the homologues encoded in the genomes of various bacteria have YedZ domains fused to transport, electron transfer and biogenesis proteins. One of the animal homologues is the 6 TMS epithelial plasma membrane antigen of the prostate (STAMP1) that is over-expressed in prostate cancer. Some animal homologues have YedZ domains fused C-terminal to homologues of NADP oxidoreductases.





