An intron is any nucleotide sequence within a gene that is removed by RNA splicing during maturation of the final RNA product. In other words, introns are non-coding regions of an RNA transcript, or the DNA encoding it, that are eliminated by splicing before translation. The word intron is derived from the term intragenic region, i.e. a region inside a gene. The term intron refers to both the DNA sequence within a gene and the corresponding sequence in RNA transcripts. Sequences that are joined together in the final mature RNA after RNA splicing are exons.

RNA splicing, in molecular biology, is a form of RNA processing in which a newly made precursor messenger RNA (pre-mRNA) transcript is transformed into a mature messenger RNA (mRNA). During splicing, introns are removed and exons are joined together. For nuclear-encoded genes, splicing takes place within the nucleus either during or immediately after transcription. For those eukaryotic genes that contain introns, splicing is usually required in order to create an mRNA molecule that can be translated into protein. For many eukaryotic introns, splicing is carried out in a series of reactions which are catalyzed by the spliceosome, a complex of small nuclear ribonucleoproteins (snRNPs). Self-splicing introns, or ribozymes capable of catalyzing their own excision from their parent RNA molecule, also exist.

Alternative splicing, or alternative RNA splicing, or differential splicing, is an alternative splicing process during gene expression that allows a single gene to code for multiple proteins. In this process, particular exons of a gene may be included within or excluded from the final, processed messenger RNA (mRNA) produced from that gene. This means the exons are joined in different combinations, leading to different (alternative) mRNA strands. Consequently, the proteins translated from alternatively spliced mRNAs will contain differences in their amino acid sequence and, often, in their biological functions. Notably, alternative splicing allows the human genome to direct the synthesis of many more proteins than would be expected from its 20,000 protein-coding genes.

A spliceosome is a large ribonucleoprotein (RNP) complex found primarily within the nucleus of eukaryotic cells. The spliceosome is assembled from small nuclear RNAs (snRNA) and numerous proteins. The spliceosome removes introns from a transcribed pre-mRNA, a type of primary transcript. This process is generally referred to as splicing. An analogy is a film editor, who selectively cuts out irrelevant or incorrect material from the initial film and sends the cleaned-up version to the director for the final cut.
The 5′ untranslated region is the region of an mRNA that is directly upstream from the initiation codon. This region is important for the regulation of translation of a transcript by differing mechanisms in viruses, prokaryotes and eukaryotes. While called untranslated, the 5′ UTR or a portion of it is sometimes translated into a protein product. This product can then regulate the translation of the main coding sequence of the mRNA. In many organisms, however, the 5′ UTR is completely untranslated, instead forming complex secondary structure to regulate translation.
The unfolded protein response (UPR) is a cellular stress response related to the endoplasmic reticulum (ER) stress. It has been found to be conserved between all mammalian species, as well as yeast and worm organisms.
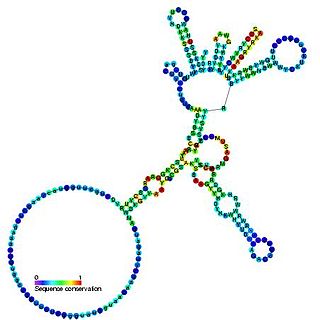
Group I introns are large self-splicing ribozymes. They catalyze their own excision from mRNA, tRNA and rRNA precursors in a wide range of organisms. The core secondary structure consists of nine paired regions (P1-P9). These fold to essentially two domains – the P4-P6 domain and the P3-P9 domain. The secondary structure mark-up for this family represents only this conserved core. Group I introns often have long open reading frames inserted in loop regions.

U6 snRNA is the non-coding small nuclear RNA (snRNA) component of U6 snRNP, an RNA-protein complex that combines with other snRNPs, unmodified pre-mRNA, and various other proteins to assemble a spliceosome, a large RNA-protein molecular complex that catalyzes the excision of introns from pre-mRNA. Splicing, or the removal of introns, is a major aspect of post-transcriptional modification and takes place only in the nucleus of eukaryotes.

TIA1 or Tia1 cytotoxic granule-associated rna binding protein is a 3'UTR mRNA binding protein that can bind the 5'TOP sequence of 5'TOP mRNAs. It is associated with programmed cell death (apoptosis) and regulates alternative splicing of the gene encoding the Fas receptor, an apoptosis-promoting protein. Under stress conditions, TIA1 localizes to cellular RNA-protein conglomerations called stress granules. It is encoded by the TIA1 gene.

X-box binding protein 1, also known as XBP1, is a protein which in humans is encoded by the XBP1 gene. The XBP1 gene is located on chromosome 22 while a closely related pseudogene has been identified and localized to chromosome 5. The XBP1 protein is a transcription factor that regulates the expression of genes important to the proper functioning of the immune system and in the cellular stress response.

DNA damage-inducible transcript 3, also known as C/EBP homologous protein (CHOP), is a pro-apoptotic transcription factor that is encoded by the DDIT3 gene. It is a member of the CCAAT/enhancer-binding protein (C/EBP) family of DNA-binding transcription factors. The protein functions as a dominant-negative inhibitor by forming heterodimers with other C/EBP members, preventing their DNA binding activity. The protein is implicated in adipogenesis and erythropoiesis, and has an important role in the cell's stress response.

The serine/threonine-protein kinase/endoribonuclease inositol-requiring enzyme 1 α (IRE1α) is an enzyme that in humans is encoded by the ERN1 gene.

The bZIP intron RNA motif is an RNA structure guiding splicing of a non-canonical intron from bZIP-containing genes called HAC1 in yeast, XBP1 in Metazoa, Hxl1 or Cib1 in Basidiomycota and bZIP60 in plants. Splicing is performed independently of the spliceosome by Ire1, a kinase with endoribonuclease activity. Exons are joined by a tRNA ligase. Recognition of the intron splice sites is mediated by a base-paired secondary structure of the mRNA that forms at the exon/intron boundaries. Splicing of the bZIP intron is a key regulatory step in the unfolded protein response (UPR). The Ire-mediated unconventional splicing was first described for HAC1 in S. cerevisiae.
Beta cells are heavily engaged in the synthesis and secretion of insulin. They are therefore particularly sensitive to endoplasmic reticulum (ER) stress and the subsequent unfolded protein response (UPR). Severe or prolonged episodes of ER stress can lead to the death of beta cells, which can contribute to the development of both Type I and Type II diabetes.

Membrane bound transcription factor peptidase, site 2 is a protein that in humans is encoded by the MBTPS2 gene.

The bZIP intron ascomycota is an unconventional bZIP intron found in some of the Ascomycota fungi, mainly in filamentous fungi from Pezizomycotina subphylum. The structure consists of two hairpins: a longer on at the 5′ and a shorter one at the 3’. Loop regions of the hairpins define the position of splice sites recognised by endoribonuclease Ire1 in response to ER stress. The unconventional splicing in this group results in excising introns of typical length 20 or 23 nt and it was first described in Trichoderma reesei and Aspergillus nidulans hacA mRNAs.

The bZIP intron basidiomycota is an unconventional bZIP intron found mainly in the Basidiomycota and some Mucoromycotina fungi. The consensus RNA structure is formed by three hairpins - two well conserved at the 5’ and 3’ ends and a variable one in between them. The loop regions of 5’ and 3’ hairpins define the splice sites recognised by Ire1, which performs the unconventional splicing in response to ER stress. In Basidiomycota, splicing results in excised introns from 20 to 101 nt in length and it was first described in Cryptococcus neoformans.

The bZIP intron plant is an unconventional bZIP intron in plants located in the mRNA of bZIP60 orthologs. The consensus RNA structure is very similar to the animal variant with short, usually 23 nt intron defined by the loop regions of the conserved hairpins. Majority of the plants contain also a nested spliceosomal intron located at the base of 3’ hairpin. The unconventional splicing in this group is performed by IRE1 in response to ER stress and it was first described in Arabidopsis thaliana.

The bZIP intron candida is an unconventional bZIP intron located in the HAC1 mRNA in a subgroup of fungi from Saccharomycetales order. So far all species with this type of structure belong to Metschnikowiaceae or Debaryomycetaceae families. However, some of the best known representatives of Debaryomycetaceae - Candida albicans and its closest relatives - contain the shorter RNA structure instead. The consensus structure consists of two well conserved hairpins with loop regions defining the unconventional splice sites. The hairpins are separated by a long insertion with conserved motifs and a predicted secondary structure. Splicing performed by Ire1 results in excision of a very long intron that was first described in Candida parapsilosis.

The bZIP intron saccharomycetales is an unconventional bZIP intron located in the HAC1 mRNA in most budding yeast belonging to Saccharomycetales order. The structure consists of two hairpins with their loop regions defining 5’ and 3’ splice sites and a long, poorly conserved sequence separating them. In some species this poorly conserved region can pair with the 5’ UTR of the HAC1 mRNA forming a pseudoknot, which stalls the translation. The unconventional splicing is performed by an endoribonuclease Ire1 in response to ER stress and it was first shown in Saccharomyces cerevisiae.
















