In biology and biochemistry, protease inhibitors, or antiproteases, are molecules that inhibit the function of proteases. Many naturally occurring protease inhibitors are proteins.
Microviridae is a family of bacteriophages with a single-stranded DNA genome. The name of this family is derived from the ancient Greek word μικρός (mikrós), meaning "small". This refers to the size of their genomes, which are among the smallest of the DNA viruses. Enterobacteria, intracellular parasitic bacteria, and spiroplasma serve as natural hosts. There are 22 species in this family, divided among seven genera and two subfamilies.

The phi X 174 bacteriophage is a single-stranded DNA (ssDNA) virus that infects Escherichia coli. This virus was isolated in 1935 by Nicolas Bulgakov in Félix d'Hérelle's laboratory at the Pasteur Institute, from samples collected in Paris sewers. Its characterization and the study of its replication mechanism were carried out from the 1950s onwards. It was the first DNA-based genome to be sequenced. This work was completed by Fred Sanger and his team in 1977. In 1962, Walter Fiers and Robert Sinsheimer had already demonstrated the physical, covalently closed circularity of ΦX174 DNA. Nobel prize winner Arthur Kornberg used ΦX174 as a model to first prove that DNA synthesized in a test tube by purified enzymes could produce all the features of a natural virus, ushering in the age of synthetic biology. In 1972–1974, Jerard Hurwitz, Sue Wickner, and Reed Wickner with collaborators identified the genes required to produce the enzymes to catalyze conversion of the single stranded form of the virus to the double stranded replicative form. In 2003, it was reported by Craig Venter's group that the genome of ΦX174 was the first to be completely assembled in vitro from synthesized oligonucleotides. The ΦX174 virus particle has also been successfully assembled in vitro. In 2012, it was shown how its highly overlapping genome can be fully decompressed and still remain functional.
Escherichia virus HK97, often shortened to HK97, is a species of virus that infects Escherichia coli and related bacteria. It is named after Hong Kong (HK), where it was first located. HK97 has a double-stranded DNA genome.
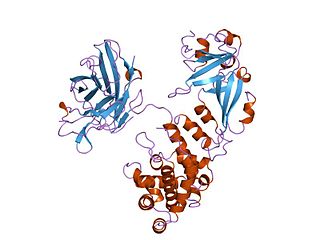
Diphtheria toxin is an exotoxin secreted mainly by Corynebacterium diphtheriae but also by Corynebacterium ulcerans and Corynebacterium pseudotuberculosis, the pathogenic bacterium that causes diphtheria. The toxin gene is encoded by a prophage called corynephage β. The toxin causes the disease in humans by gaining entry into the cell cytoplasm and inhibiting protein synthesis.
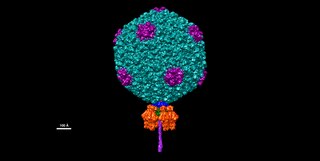
Salmonella virus P22 is a bacteriophage in the Podoviridae family that infects Salmonella typhimurium. Like many phages, it has been used in molecular biology to induce mutations in cultured bacteria and to introduce foreign genetic material. P22 has been used in generalized transduction and is an important tool for investigating Salmonella genetics.

A leucine-rich repeat (LRR) is a protein structural motif that forms an α/β horseshoe fold. It is composed of repeating 20–30 amino acid stretches that are unusually rich in the hydrophobic amino acid leucine. These tandem repeats commonly fold together to form a solenoid protein domain, termed leucine-rich repeat domain. Typically, each repeat unit has beta strand-turn-alpha helix structure, and the assembled domain, composed of many such repeats, has a horseshoe shape with an interior parallel beta sheet and an exterior array of helices. One face of the beta sheet and one side of the helix array are exposed to solvent and are therefore dominated by hydrophilic residues. The region between the helices and sheets is the protein's hydrophobic core and is tightly sterically packed with leucine residues.

Carbamoyl phosphate synthetase catalyzes the ATP-dependent synthesis of carbamoyl phosphate from glutamine or ammonia and bicarbonate. This enzyme catalyzes the reaction of ATP and bicarbonate to produce carboxy phosphate and ADP. Carboxy phosphate reacts with ammonia to give carbamic acid. In turn, carbamic acid reacts with a second ATP to give carbamoyl phosphate plus ADP.
HHV Capsid Portal Protein, or HSV-1 UL-6 protein, is the protein which forms a cylindrical portal in the capsid of Herpes simplex virus (HSV-1). The protein is commonly referred to as the HSV-1 UL-6 protein because it is the transcription product of Herpes gene UL-6.
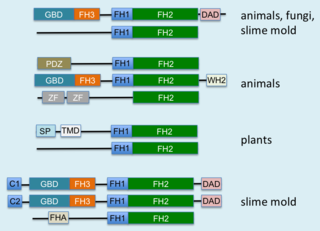
Formins (formin homology proteins) are a group of proteins that are involved in the polymerization of actin and associate with the fast-growing end (barbed end) of actin filaments. Most formins are Rho-GTPase effector proteins. Formins regulate the actin and microtubule cytoskeleton and are involved in various cellular functions such as cell polarity, cytokinesis, cell migration and SRF transcriptional activity. Formins are multidomain proteins that interact with diverse signalling molecules and cytoskeletal proteins, although some formins have been assigned functions within the nucleus.

Cullins are a family of hydrophobic scaffold proteins which provide support for ubiquitin ligases (E3). All eukaryotes appear to have cullins. They combine with RING proteins to form Cullin-RING ubiquitin ligases (CRLs) that are highly diverse and play a role in myriad cellular processes, most notably protein degradation by ubiquitination.
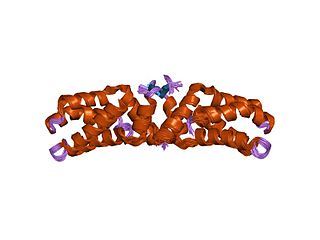
The L27 domain is a protein domain that is found in receptor targeting proteins Lin-2 and Lin-7, as well as some protein kinases and human MPP2 protein. The L27 domain is a protein interaction module that exists in a large family of scaffold proteins, functioning as an organisation centre of large protein assemblies required for the establishment and maintenance of cell polarity. L27 domains form specific heterotetrameric complexes, in which each domain contains three alpha-helices. The L27_2 domain is a protein-protein interaction domain capable of organising scaffold proteins into supramolecular assemblies by formation of heteromeric L27_2 domain complexes. L27_2 domain-mediated protein assemblies have been shown to play essential roles in cellular processes including asymmetric cell division, establishment and maintenance of cell polarity, and clustering of receptors and ion channels. Members of this family form specific heterotetrameric complexes, in which each domain contains three alpha-helices. The two N-terminal helices of each L27_2 domain pack together to form a tight, four-helix bundle in the heterodimer, whilst the third helix of each L27_2 domain forms another four-helix bundle that assembles the two units of the heterodimer into a tetramer.

In molecular biology, the hexon protein is a major coat protein found in adenoviruses. Hexon coat proteins are synthesised during late infection and form homo-trimers. The 240 copies of the hexon trimer that are produced are organised so that 12 lie on each of the 20 facets. The central 9 hexons in a facet are cemented together by 12 copies of polypeptide IX. The penton complex, formed by the peripentonal hexons and penton base, lie at each of the 12 vertices. The hexon coat protein is a duplication consisting of two domains with a similar fold packed together like the nucleoplasmin subunits. Within a hexon trimer, the domains are arranged around a pseudo 6-fold axis. The domains have a beta-sandwich structure consisting of 8 strands in two sheets with a jelly-roll topology; each domain is heavily decorated with many insertions. Some hexon proteins contain a distinct C-terminal domain.
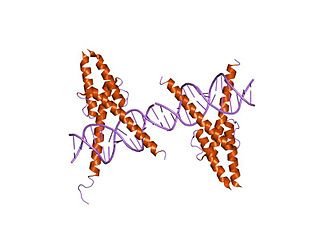
In molecular biology, the myogenic determination factor 5 proteins are a family of proteins found in eukaryotes. This family includes the Myf5 protein, which is responsible for directing cells to the skeletal myocyte lineage during development. Myf5 is likely to act in a similar way to the other MRF4 proteins such as MyoD which perform the same function. These are histone acetyltransferases and histone deacetylases which activate and repress genes involved in the myocyte lineage.

In molecular biology, a carbohydrate-binding module (CBM) is a protein domain found in carbohydrate-active enzymes. The majority of these domains have carbohydrate-binding activity. Some of these domains are found on cellulosomal scaffoldin proteins. CBMs were previously known as cellulose-binding domains. CBMs are classified into numerous families, based on amino acid sequence similarity. There are currently 64 families of CBM in the CAZy database.

In molecular biology, the cohesin domain is a protein domain. It interacts with a complementary domain, termed the dockerin domain. The cohesin-dockerin interaction is the crucial interaction for complex formation in the cellulosome.

In molecular biology, the Cro repressor family is a family of repressor proteins in bacteriophage lambda that includes the Cro repressor.

In molecular biology, excisionase is a bacteriophage protein encoded by the Xis gene. It is involved in excisive recombination by regulating the assembly of the excisive intasome and by inhibiting viral integration. It adopts an unusual winged-helix structure in which two alpha helices are packed against two extended strands. Also present in the structure is a two-stranded anti-parallel beta-sheet, whose strands are connected by a four-residue wing. During interaction with DNA, helix alpha2 is thought to insert into the major groove, while the wing contacts the adjacent minor groove or phosphodiester backbone. The C-terminal region of excisionase is involved in interaction with phage-encoded integrase (Int), and a putative C-terminal alpha helix may fold upon interaction with Int and/or DNA.

In molecular biology, a phage major coat protein is an alpha-helical protein that forms a viral envelope of filamentous bacteriophages. These bacteriophages are flexible rods, about one to two micrometres long and six nm in diameter, with a helical shell of protein subunits surrounding a DNA core. The approximately 50-residue subunit of the major coat protein is largely alpha-helix, and the axis of the alpha-helix makes a small angle with the axis of the virion. The protein shell can be considered in three sections: the outer surface, occupied by the N-terminal region of the subunit and rich in acidic residues that give the virion a low isoelectric point; the interior of the shell where protein subunits interact, mainly with each other; and the inner surface, rich in positively charged residues that interact with the DNA core.

In molecular biology, the YqaJ refers to the YqaJ/K domain from the skin prophage of the bacterium, Bacillus subtilis. This protein domain, often found in bacterial species, is actually of viral origin. The protein forms an oligomer and functions as an alkaline exonuclease, or in simpler terms, an enzyme that digests double-stranded DNA. It is a reaction which is dependent on Magnesium. It has a preference for 5'-phosphorylated DNA ends. It thus forms part of the two-component SynExo viral recombinase functional unit.

















