A regulatory sequence is a segment of a nucleic acid molecule which is capable of increasing or decreasing the expression of specific genes within an organism. Regulation of gene expression is an essential feature of all living organisms and viruses.
A satellite is a subviral agent composed of nucleic acid that depends on the co-infection of a host cell with a helper virus for its replication.

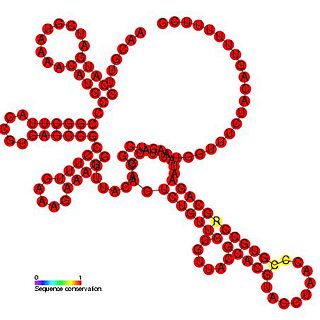
The Citrus tristeza virus replication signal is a regulatory element involved in a viral replication signal which is highly conserved in citrus tristeza viruses. Replication signals are required for viral replication and are usually found near the 5' and 3' termini of protein coding genes. This element is predicted to form ten stem loop structures some of which are essential for functions that provide for efficient viral replication.
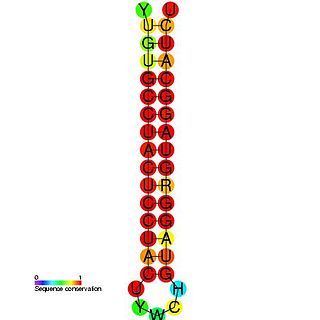
The Hepatitis C stem-loop IV is part of a putative RNA element found in the NS5B coding region. This element along with stem-loop VII, is important for colony formation, though its exact function and mechanism are unknown.

The Hepatitis C virus (HCV) cis-acting replication element (CRE) is an RNA element which is found in the coding region of the RNA-dependent RNA polymerase NS5B. Mutations in this family have been found to cause a blockage in RNA replication and it is thought that both the primary sequence and the structure of this element are crucial for HCV RNA replication.
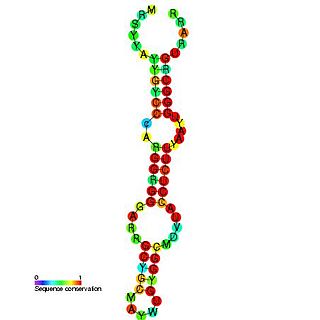
Hepatitis C virus stem-loop VII is a regulatory element found in the coding region of the RNA-dependent RNA polymerase gene, NS5B. Similarly to stem-loop IV, the stem-loop structure is important for colony formation, though its exact function and mechanism are unknown.
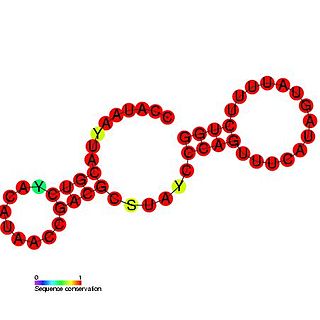
The Rubella virus 3' cis-acting element RNA family represents a cis-acting element found at the 3' UTR in the rubella virus. This family contains three conserved step loop structures. Calreticulin (CAL), which is known to bind calcium in most eukaryotic cells, is able to specifically bind to the first stem loop of this RNA. CAL binding is thought to be related to viral pathogenesis and in particular arthritis which occurs frequently in rubella infections in adults and is independent of viral viability. All stem loop structures are thought to be important for efficient viral replication and deletion of stem loop three is known to be lethal.
The Togavirus 5' plus strand cis-regulatory element is an RNA element which is thought to be essential for both plus and minus strand RNA synthesis.
Tombus virus defective interfering (DI) RNA region 3 is an important cis-regulatory region identified in the 3' UTR of Tombusvirus defective interfering particles (DI).

The Upstream pseudoknot (UPSK) domain is an RNA element found in the turnip yellow mosaic virus, beet virus Q, barley stripe mosaic virus and tobacco mosaic virus, which is thought to be needed for efficient transcription. Disruption of the pseudoknot structure gives rise to a 50% drop in transcription efficiency. This element acts in conjunction with the Tymovirus/Pomovirus tRNA-like 3' UTR element to enhance translation.
Potexvirus is a genus of pathogenic viruses in the order Tymovirales, in the family Alphaflexiviridae. Plants serve as natural hosts. There are currently 37 species in this genus including the type species Potato virus X. Diseases associated with this genus include: mosaic and ringspot symptoms. The genus name comes from the type species POTato Virus X).
Alphaflexiviridae is a family of viruses in the order Tymovirales. Plants and fungi serve as natural hosts. There are currently 51 species in this family, divided among 6 genera. Diseases associated with this family include: mosaic and ringspot symptoms.
Cis-acting replication elements bring together the 5' and 3' ends during replication of positive-sense single-stranded RNA viruses and double-stranded RNA viruses.
In molecular biology, the Norovirus cis-acting replication element (CRE) is an RNA element which is found in the coding region of the RNA-dependent RNA polymerase in Norovirus. It occurs near to the 5' end of the RNA dependant RNA polymerase gene, this is the same location that the Hepatitis A virus cis-acting replication element is found in.