In biology and biochemistry, protease inhibitors, or antiproteases, are molecules that inhibit the function of proteases. Many naturally occurring protease inhibitors are proteins.
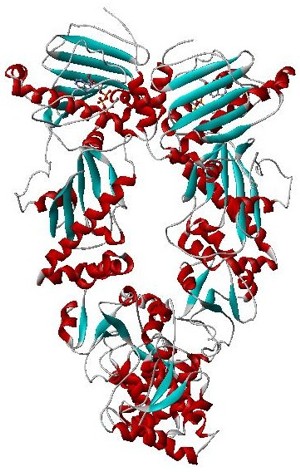
Hsp90 is a chaperone protein that assists other proteins to fold properly, stabilizes proteins against heat stress, and aids in protein degradation. It also stabilizes a number of proteins required for tumor growth, which is why Hsp90 inhibitors are investigated as anti-cancer drugs.
AAAproteins are a large group of protein family sharing a common conserved module of approximately 230 amino acid residues. This is a large, functionally diverse protein family belonging to the AAA+ protein superfamily of ring-shaped P-loop NTPases, which exert their activity through the energy-dependent remodeling or translocation of macromolecules.
Macromolecular docking is the computational modelling of the quaternary structure of complexes formed by two or more interacting biological macromolecules. Protein–protein complexes are the most commonly attempted targets of such modelling, followed by protein–nucleic acid complexes.
Anthrax toxin is a three-protein exotoxin secreted by virulent strains of the bacterium, Bacillus anthracis—the causative agent of anthrax. The toxin was first discovered by Harry Smith in 1954. Anthrax toxin is composed of a cell-binding protein, known as protective antigen (PA), and two enzyme components, called edema factor (EF) and lethal factor (LF). These three protein components act together to impart their physiological effects. Assembled complexes containing the toxin components are endocytosed. In the endosome, the enzymatic components of the toxin translocate into the cytoplasm of a target cell. Once in the cytosol, the enzymatic components of the toxin disrupt various immune cell functions, namely cellular signaling and cell migration. The toxin may even induce cell lysis, as is observed for macrophage cells. Anthrax toxin allows the bacteria to evade the immune system, proliferate, and ultimately kill the host animal. Research on anthrax toxin also provides insight into the generation of macromolecular assemblies, and on protein translocation, pore formation, endocytosis, and other biochemical processes.

A leucine-rich repeat (LRR) is a protein structural motif that forms an α/β horseshoe fold. It is composed of repeating 20–30 amino acid stretches that are unusually rich in the hydrophobic amino acid leucine. These tandem repeats commonly fold together to form a solenoid protein domain, termed leucine-rich repeat domain. Typically, each repeat unit has beta strand-turn-alpha helix structure, and the assembled domain, composed of many such repeats, has a horseshoe shape with an interior parallel beta sheet and an exterior array of helices. One face of the beta sheet and one side of the helix array are exposed to solvent and are therefore dominated by hydrophilic residues. The region between the helices and sheets is the protein's hydrophobic core and is tightly sterically packed with leucine residues.

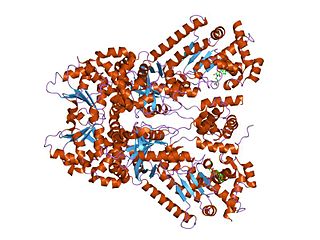
EF-Tu is a prokaryotic elongation factor responsible for catalyzing the binding of an aminoacyl-tRNA (aa-tRNA) to the ribosome. It is a G-protein, and facilitates the selection and binding of an aa-tRNA to the A-site of the ribosome. As a reflection of its crucial role in translation, EF-Tu is one of the most abundant and highly conserved proteins in prokaryotes. It is found in eukaryotic mitochondria as TUFM.

Axin-1 is a protein that in humans is encoded by the AXIN1 gene.

Engulfment and cell motility protein 1 is a protein that in humans is encoded by the ELMO1 gene. ELMO1 is located on chromosome number seven in humans and is located on chromosome number thirteen in mice.

The inhibitor of apoptosis domain -- also known as IAP repeat, Baculovirus Inhibitor of apoptosis protein Repeat, or BIR -- is a structural motif found in proteins with roles in apoptosis, cytokine production, and chromosome segregation. Proteins containing BIR are known as inhibitor of apoptosis proteins (IAPs), or BIR-containing proteins, and include BIRC1 (NAIP), BIRC2 (cIAP1), BIRC3 (cIAP2), BIRC4 (xIAP), BIRC5 (survivin) and BIRC6.

In molecular biology, phosphotyrosine-binding domains are protein domains which bind to phosphotyrosine.
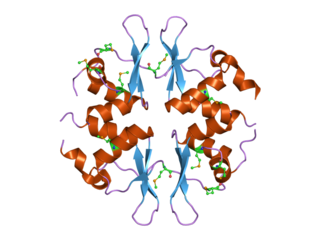
In molecular biology, the CBS domain is a protein domain found in a range of proteins in all species from bacteria to humans. It was first identified as a conserved sequence region in 1997 and named after cystathionine beta synthase, one of the proteins it is found in. CBS domains are also found in a wide variety of other proteins such as inosine monophosphate dehydrogenase, voltage gated chloride channels and AMP-activated protein kinase (AMPK). CBS domains regulate the activity of associated enzymatic and transporter domains in response to binding molecules with adenosyl groups such as AMP and ATP, or s-adenosylmethionine.

Monobodies are synthetic binding proteins constructed using a fibronectin type III domain (FN3) as a molecular scaffold. Specifically, this class of binding proteins are built upon a diversified library of the 10th FN3 domain of human fibronectin. Monobodies are a simple and robust alternative to antibodies for creating target-binding proteins. The hybrid term monobody was coined in 1998 by the Koide group who published the first paper demonstrating the monobody concept using the tenth FN3 domain of human fibronectin.

The GHKL domain is an evolutionary conserved protein domain. It is an ATPase domain found in several ATP-binding proteins such as histidine kinase, DNA gyrase B, topoisomerases, heat shock protein HSP90, phytochrome-like ATPases and DNA mismatch repair proteins.

BIM-1 and the related compounds BIM-2, BIM-3, and BIM-8 are bisindolylmaleimide-based protein kinase C (PKC) inhibitors. These inhibitors also inhibit PDK1 explaining the higher inhibitory potential of LY33331 compared to the other BIM compounds a bisindolylmaleimide inhibitor toward PDK1.

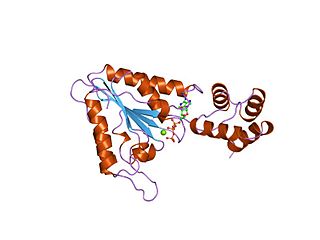
Ras association domain-containing protein 9 (RASSF9), also known as PAM COOH-terminal interactor protein 1 (PCIP1) or peptidylglycine alpha-amidating monooxygenase COOH-terminal interactor (PAMCI) is a protein that in humans is encoded by the RASSF9 gene.
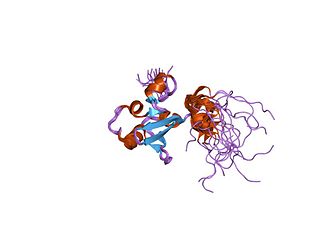
In molecular biology, the protein domain SAND is named after a range of proteins in the protein family: Sp100, AIRE-1, NucP41/75, DEAF-1. It is localised in the cell nucleus and has an important function in chromatin-dependent transcriptional control. It is found solely in eukaryotes.
WH1 domain is an evolutionary conserved protein domain found on WASP proteins, which are often involved in actin polymerization.
Amide Rings are small motifs in proteins and polypeptides. They consist of 9-atom or 11-atom rings formed by two CO...HN hydrogen bonds between a side chain amide group and the main chain atoms of a short polypeptide. They are observed with glutamine or asparagine side chains within proteins and polypeptides. Structurally similar rings occur in the binding of purine, pyrimidine and nicotinamide bases to the main chain atoms of proteins. About 4% of asparagines and glutamines form amide rings; in databases of protein domain structures, one is present, on average, every other protein.

Natalie C. J. Strynadka FRS is a professor of Biochemistry in the Department of Biochemistry and Molecular Biology at the University of British Columbia.

















