Related Research Articles
Ebbe Schmidt Nielsen was a Danish entomologist influential in systematics and Lepidoptera research, and an early proponent of biodiversity informatics. The journal Invertebrate Systematics was established with significant contributions from Nielsen, and he assisted in the founding of the Global Biodiversity Information Facility (GBIF). Nielsen wrote several books, published over eighty scientific papers, and was highly regarded within the scientific community. Following his death, the GBIF organised the Ebbe Nielsen Prize in his memory, awarded annually to promising researchers in the field of biodiversity informatics. The moth Pollanisus nielseni is named after Nielsen.

Calathus is a genus of ground beetle native to the Palearctic, the Near East and North Africa. There are at least 190 described species in Calathus.
Biodiversity informatics is the application of informatics techniques to biodiversity information, such as taxonomy, biogeography or ecology. It is defined as the application of Information technology technologies to management, algorithmic exploration, analysis and interpretation of primary data regarding life, particularly at the species level organization. Modern computer techniques can yield new ways to view and analyze existing information, as well as predict future situations. Biodiversity informatics is a term that was only coined around 1992 but with rapidly increasing data sets has become useful in numerous studies and applications, such as the construction of taxonomic databases or geographic information systems. Biodiversity informatics contrasts with "bioinformatics", which is often used synonymously with the computerized handling of data in the specialized area of molecular biology.

Apache Taverna was an open source software tool for designing and executing workflows, initially created by the myGrid project under the name Taverna Workbench, then a project under the Apache incubator. Taverna allowed users to integrate many different software components, including WSDL SOAP or REST Web services, such as those provided by the National Center for Biotechnology Information, the European Bioinformatics Institute, the DNA Databank of Japan (DDBJ), SoapLab, BioMOBY and EMBOSS. The set of available services was not finite and users could import new service descriptions into the Taverna Workbench.

The Encyclopedia of Life (EOL) is a free, online encyclopedia intended to document all of the 1.9 million living species known to science. It aggregates content to form "page"s for every known species. Content is compiled from existing trusted databases which are curated by experts and it calls on the assistance of non-experts throughout the world. It includesvideo, sound, images, graphics, information on characteristics, as well as text.. In addition, the Encyclopedia incorporates species-related content from the Biodiversity Heritage Library, which digitizes millions of pages of printed literature from the world's major natural history libraries. The BHL digital content is indexed with the names of organisms using taxonomic indexing software developed by the Global Names project. The EOL project was initially backed by a US$50 million funding commitment, led by the MacArthur Foundation and the Sloan Foundation, who provided US$20 million and US$5 million, respectively. The additional US$25 million came from five cornerstone institutions—the Field Museum, Harvard University, the Marine Biological Laboratory, the Missouri Botanical Garden, and the Smithsonian Institution. The project was initially led by Jim Edwards and the development team by David Patterson. Today, participating institutions and individual donors continue to support EOL through financial contributions.
Darwin Core is an extension of Dublin Core for biodiversity informatics. It is meant to provide a stable standard reference for sharing information on biological diversity (biodiversity). The terms described in this standard are a part of a larger set of vocabularies and technical specifications under development and maintained by Biodiversity Information Standards (TDWG).
A taxonomic database is a database created to hold information on biological taxa – for example groups of organisms organized by species name or other taxonomic identifier – for efficient data management and information retrieval. Taxonomic databases are routinely used for the automated construction of biological checklists such as floras and faunas, both for print publication and online; to underpin the operation of web-based species information systems; as a part of biological collection management ; as well as providing, in some cases, the taxon management component of broader science or biology information systems. They are also a fundamental contribution to the discipline of biodiversity informatics.
Plazi is a Swiss-based international non-profit association supporting and promoting the development of persistent and openly accessible digital bio-taxonomic literature. Plazi is cofounder of the Biodiversity Literature Repository and is maintaining this digital taxonomic literature repository at Zenodo to provide access to FAIR data converted from taxonomic publications using the TreatmentBank service, enhances submitted taxonomic treatments by creating a version in the XML format Taxpub, and educates about the importance of maintaining open access to scientific discourse and data. It is a contributor to the evolving e-taxonomy in the field of Biodiversity Informatics.

DataONE is a network of interoperable data repositories facilitating data sharing, data discovery, and open science. Originally supported by $21.2 million in funding from the US National Science Foundation as one of the initial DataNet programs in 2009, funding was renewed in 2014 through 2020 with an additional $15 million. DataONE helps preserve, access, use, and reuse of multi-discipline scientific data through the construction of primary cyberinfrastructure and an education and outreach program. DataONE provides scientific data archiving for ecological and environmental data produced by scientists. DataONE's goal is to preserve and provide access to multi-scale, multi-discipline, and multi-national data. Users include scientists, ecosystem managers, policy makers, students, educators, librarians, and the public.
Figshare is an online open access repository where researchers can preserve and share their research outputs, including figures, datasets, images, and videos. It is free to upload content and free to access, in adherence to the principle of open data. Figshare is one of a number of portfolio businesses supported by Digital Science, a subsidiary of Springer Nature.
Pensoft Publishers are a publisher of scientific literature based in Sofia, Bulgaria. Pensoft was founded in 1992, by two academics: Lyubomir Penev and Sergei Golovatch. It has published over 1000 academic and professional books and currently publishes over 60 peer-reviewed open access scientific journals including ZooKeys, PhytoKeys, Check List, Comparative Cytogenetics, Journal of Hymenoptera Research, Deutsche Entomologische Zeitschrift, and Zoosystematics and Evolution.

Zenodo is a general-purpose open repository developed under the European OpenAIRE program and operated by CERN. It allows researchers to deposit research papers, data sets, research software, reports, and any other research related digital artefacts. For each submission, a persistent digital object identifier (DOI) is minted, which makes the stored items easily citeable.

Huechys phaenicura is a species of cicada belonging to the family Cicadidae.

In Belgium, open access to scholarly communication accelerated after 2007 when the University of Liège adopted its first open-access mandate. The "Brussels Declaration" for open access was signed by officials in 2012.

In India, Open Access movement started in May 2004, when two workshops were organized by the M S Swaminathan Research Foundation, Chennai. In 2006, the National Knowledge Commission in its recommendations proposed that "access to knowledge is the most fundamental way of increasing the opportunities and reach of individuals and groups". In 2009, the Council of Scientific & Industrial Research (CSIR) began requiring that its grantees provide open access to funded research. In 2011, the Open Access India forum formulated a draft policy on Open Access for India. Shodhganga, a digital repository for theses, was established in 2011 with the aim of promoting and preserving academic research. The University Grants Commission (UGC) made it mandatory for scholars to deposit their theses in Shodhganga, as per the Minimum Standards and Procedure for Award of M. Phil./Ph.D. Degrees Regulations, 2016.Currently, the Directory of Open Access Journals lists 326 open access journals published in India, of which 233 have no fees.

FAIR data are data which meet principles of findability, accessibility, interoperability, and reusability (FAIR). The acronym and principles were defined in a March 2016 paper in the journal Scientific Data by a consortium of scientists and organizations.

The Interim Register of Marine and Nonmarine Genera (IRMNG) is a taxonomic database which attempts to cover published genus names for all domains of life, from 1758 in zoology up to the present, arranged in a single, internally consistent taxonomic hierarchy, for the benefit of Biodiversity Informatics initiatives plus general users of biodiversity (taxonomic) information. In addition to containing just over 500,000 published genus name instances as at May 2023, the database holds over 1.7 million species names, although this component of the data is not maintained in as current or complete state as the genus-level holdings. IRMNG can be queried online for access to the latest version of the dataset and is also made available as periodic snapshots or data dumps for import/upload into other systems as desired. The database was commenced in 2006 at the then CSIRO Division of Marine and Atmospheric Research in Australia and, since 2016, has been hosted at the Flanders Marine Institute (VLIZ) in Belgium.
The Ebbe Nielsen Challenge is an international science competition conducted annually from 2015 onwards by the Global Biodiversity Information Facility (GBIF), with a set of cash prizes that recognize researcher(s)' submissions in creating software or approaches that successfully address a GBIF-issued challenge in the field of biodiversity informatics. It succeeds the Ebbe Nielsen Prize, which was awarded annually by GBIF between 2002 and 2014. The name of the challenge honours the memory of prominent entomologist and biodiversity informatics proponent Ebbe Nielsen, who died of a heart attack in the U.S.A. en route to the 2001 GBIF Governing Board meeting.

Gregor Hagedorn is a German botanist and academic director at the Natural History Museum, Berlin.
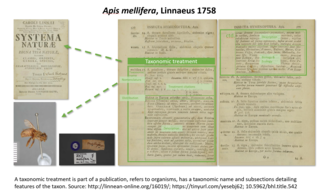
A taxonomic treatment is a section in a scientific publication documenting the features of a related group of organisms or taxa. Treatments have been the building blocks of how data about taxa are provided, ever since the beginning of modern taxonomy by Linnaeus 1753 for plants and 1758 for animals. Each scientifically described taxon has at least one taxonomic treatment. In today’s publishing, a taxonomic treatment tag
References
- ↑ "Biodiversity Literature Repository". zenodo.org/communities/biosyslit/. Retrieved 2021-04-25.
- ↑ "Zenodo's Biodiversity Literature Repository is a treasure-trove of FAIR data". www.openaire.eu/. Retrieved 2021-04-25.
- ↑ "Taxonomic treatments in BLR". zenodo.org/communities/biosyslit/. Retrieved 2021-04-25.
- ↑ "Figures in BLR". zenodo.org/communities/biosyslit/. Retrieved 2021-04-25.
- ↑ "Articles in BLR". zenodo.org/communities/biosyslit/. Retrieved 2021-04-25.
- ↑ "TreatmentBank". plazi.org. Archived from the original on 2021-05-29. Retrieved 2021-04-25.
- ↑ Agosti, Donat; Guidoti, Marcus; Sautter, Guido (2020). "Liberating the Richness of Facts implicit in taxonomic Publication: The Plazi Workflow". Biodiversity Information Science and Standards. 4 (e59179). doi: 10.3897/biss.4.59179 .
- ↑ Agosti, Donat; Egloff, Willi (2008). "Taxonomic information exchange and copyright: the Plazi approach". BMC Research Notes. 2 (53). doi: 10.1186/1756-0500-2-53 . PMC 2673227 .
- ↑ "Plazi.org taxonomic treatments database". gbif.org. Retrieved 2021-04-25.