Related Research Articles
Biomedical text mining refers to the methods and study of how text mining may be applied to texts and literature of the biomedical and molecular biology domains. As a field of research, biomedical text mining incorporates ideas from natural language processing, bioinformatics, medical informatics and computational linguistics. The strategies developed through studies in this field are frequently applied to the biomedical and molecular biology literature available through services such as PubMed.

The Global Biodiversity Information Facility (GBIF) is an international organisation that focuses on making scientific data on biodiversity available via the Internet using web services. The data are provided by many institutions from around the world; GBIF's information architecture makes these data accessible and searchable through a single portal. Data available through the GBIF portal are primarily distribution data on plants, animals, fungi, and microbes for the world, and scientific names data.
The Clinical Data Interchange Standards Consortium (CDISC) is a standards developing organization (SDO) dealing with medical research data linked with healthcare, to "enable information system interoperability to improve medical research and related areas of healthcare". The standards support medical research from protocol through analysis and reporting of results and have been shown to decrease resources needed by 60% overall and 70–90% in the start-up stages when they are implemented at the beginning of the research process.
Biodiversity informatics is the application of informatics techniques to biodiversity information, such as taxonomy, biogeography or ecology. Modern computer techniques can yield new ways to view and analyze existing information, as well as predict future situations. Biodiversity informatics is a term that was only coined around 1992 but with rapidly increasing data sets has become useful in numerous studies and applications, such as the construction of taxonomic databases or geographic information systems. Biodiversity informatics contrasts with "bioinformatics", which is often used synonymously with the computerized handling of data in the specialized area of molecular biology.
The National Centre for Text Mining (NaCTeM) is a publicly funded text mining (TM) centre. It was established to provide support, advice, and information on TM technologies and to disseminate information from the larger TM community, while also providing tailored services and tools in response to the requirements of the United Kingdom academic community.
The Semantic Interoperability Centre Europe (SEMIC.EU) was an eGovernment service initiated by the European Commission and managed by the Interoperable Delivery of European eGovernment Services to public Administrations, Businesses and Citizens (IDABC) Unit. As one of the 'horizontal measures' of the IDABC, it was established as a permanent implementation of the principles stipulated in the 'European Interoperability Framework' (EIF). It offered a service for the exchange of semantic interoperability solutions, with a focus on demands of eGovernment in Europe. Through the establishment of a single sharing and collaboration point, the European Union wanted to resolve the problems of semantic interoperability amongst the EU member states. The main idea behind the service was to make visible specifications that already exist, so as to increase their reuse. In this way, governmental agencies and developers benefit as they do not reinvent the wheel, they reduce development costs, and increase the interoperability of their systems.
Darwin Core is an extension of Dublin Core for biodiversity informatics. It is meant to provide a stable standard reference for sharing information on biological diversity (biodiversity). The terms described in this standard are a part of a larger set of vocabularies and technical specifications under development and maintained by Biodiversity Information Standards (TDWG).
PhyloXML is an XML language for the analysis, exchange, and storage of phylogenetic trees and associated data. The structure of phyloXML is described by XML Schema Definition (XSD) language.
Computational Resources for Drug Discovery (CRDD) is one of the important silico modules of Open Source for Drug Discovery (OSDD). The CRDD web portal provides computer resources related to drug discovery on a single platform. It provides computational resources for researchers in computer-aided drug design, a discussion forum, and resources to maintain Wikipedia related to drug discovery, predict inhibitors, and predict the ADME-Tox property of molecules One of the major objectives of CRDD is to promote open source software in the field of chemoinformatics and pharmacoinformatics.
Darwin Core Archive (DwC-A) is a biodiversity informatics data standard that makes use of the Darwin Core terms to produce a single, self-contained dataset for species occurrence, checklist, sampling event or material sample data. Essentially it is a set of text (CSV) files with a simple descriptor (meta.xml) to inform others how your files are organized. The format is defined in the Darwin Core Text Guidelines. It is the preferred format for publishing data to the GBIF network.
Data publishing is the act of releasing research data in published form for use by others. It is a practice consisting in preparing certain data or data set(s) for public use thus to make them available to everyone to use as they wish. This practice is an integral part of the open science movement. There is a large and multidisciplinary consensus on the benefits resulting from this practice.
Pensoft Publishers are a publisher of scientific literature based in Sofia, Bulgaria. Pensoft was founded in 1992, by two academics: Lyubomir Penev and Sergei Golovatch. It has published over 1000 academic and professional books and currently publishes over 60 peer-reviewed open access scientific journals including ZooKeys, PhytoKeys, Check List, Comparative Cytogenetics, Journal of Hymenoptera Research, Deutsche Entomologische Zeitschrift, and Zoosystematics and Evolution.

Catherine Norton was an American librarian. She was the first Director of Information Systems at the Marine Biological Laboratory (MBL).

The Interim Register of Marine and Nonmarine Genera (IRMNG) is a taxonomic database which attempts to cover published genus names for all domains of life from 1758 in zoology up to the present, arranged in a single, internally consistent taxonomic hierarchy, for the benefit of Biodiversity Informatics initiatives plus general users of biodiversity (taxonomic) information. In addition to containing over 490,000 published genus name instances as at March 2020, the database holds over 1.7 million species names, although this component of the data is not maintained in as current or complete state as the genus-level holdings. IRMNG can be queried online for access to the latest version of the dataset and is also made available as periodic snapshots or data dumps for import/upload into other systems as desired.
The Ebbe Nielsen Challenge is an international science competition conducted annually from 2015 onwards by the Global Biodiversity Information Facility (GBIF), with a set of cash prizes that recognize researcher(s)' submissions in creating software or approaches that successfully address a GBIF-issued challenge in the field of biodiversity informatics. It succeeds the Ebbe Nielsen Prize, which was awarded annually by GBIF between 2002 and 2014. The name of the challenge honours the memory of prominent entomologist and biodiversity informatics proponent Ebbe Nielsen, who died of a heart attack in the U.S.A. en route to the 2001 GBIF Governing Board meeting.

Nomenclator Zoologicus is one of the major compendia in the field of zoological nomenclature, compiled by Sheffield Airey Neave and his successors and published in 9 volumes over the period 1939–1994, under the auspices of the Zoological Society of London; a tenth, electronic-only volume was also produced before the project ceased. It contains over 340,000 published name instances with their authorities and details of their original publication, certain nomenclatural notes and cross references, and an indication of the taxonomic group to which each is assigned. An electronic (digitised) version of volumes 1-10 was released online by the uBio project, based at the Marine Biological Laboratory, Woods Hole, in 2004–2005.

Anthony J. J. ("Tony") Rees is a British-born software developer, data manager and biologist resident in Australia since 1986, and previously a data manager with CSIRO Marine and Atmospheric Research. He is responsible for developing a number of software systems currently used in science data management, including c-squares, Taxamatch, and IRMNG, the Interim Register of Marine and Nonmarine Genera. He has also been closely involved with the development of other biodiversity informatics initiatives including the Ocean Biogeographic Information System (OBIS), AquaMaps, and the iPlant Taxonomic Name Resolution Service (TNRS).
The Biodiversity Literature Repository (BLR) is a biodiversity dedicated community created in November 11, 2013, in Zenodo, the open science repository at CERN and part of the European project OpenAIRE. The goal of BLR is to provide a long-term, stable, open repository that allows deposition of bio-taxonomic articles enhanced with custom metadata and links to data extracted from therein and deposited in BLR. As of April 25, 2021, this includes 94,443 taxonomic treatments and 293,457 figures from 48,993 articles which are made findable, accessible, interoperable and reusable FAIR data. Most of the data is uploaded on a continuous basis by Plazi using its TreatmentBank service based on their Plazi workflow, and Pensoft Publishers using BLR as repository for data published in their journals. The largest single re-user of data is the Global Biodiversity Information Facility (GBIF), using data from within 33,623 processed articles.
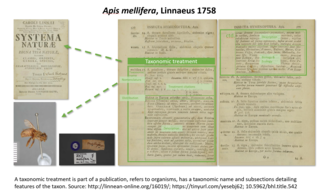
Taxonomic treatment refers to a section in a scientific publication documenting the features of a related group of organisms or taxa. Treatments have been the building blocks of how data about taxa are provided, ever since the beginning of modern taxonomy by Linnaeus 1753 for plants and 1758 for animals. Each scientifically described taxon has at least one taxonomic treatment. In today’s publishing, a taxonomic treatment tag is used to delimit such a section. It allows to make this section findable, accessible, interoperable and reusable FAIR data. This is implemented in the Biodiversity Literature Repository, where upon deposition of the treatment a persistent DataCite digital object identifier (DOI) is minted. This includes metadata about the treatment, the source publication and other cited resources, such as figures cited in the treatment. This DOI allows a link from a taxonomic name usage to the respective scientific evidence provided by the author(s), both for human and machine consumption.
References
- ↑ "TreatmentBank". plazi.org. Retrieved 2020-02-09.
- ↑ Catapano, Terry. "Taxpub". Plazi. Retrieved 27 October 2009.
- ↑ Zauner, H (2009). "Evolving e-taxonomy". BMC Evolutionary Biology. 9: 141. doi:10.1186/1471-2148-9-141. PMC 2714837 . PMID 19555493.
- ↑ Sautter, Guido. "GoldenGATE Editor". Plazi, Technische Hochschule Karlsruhe. Retrieved 27 October 2009.
- ↑ "Plazidata in GBIF". Plazi. Archived from the original on 3 March 2016. Retrieved 27 October 2009.
- ↑ "Plazi.org taxonomic treatments database". gbif.org. Retrieved 2021-04-25.
- ↑ Agosti D, Egloff W (2009). "Taxonomic information exchange and copyright: the Plazi approach". BMC Research Notes. 2: 53. doi:10.1186/1756-0500-2-53. PMC 2673227 . PMID 19331688.
- ↑ Costello M J (2009). "Motivating Online Publication of Data". BioScience. 59:5 (5): 418–427. doi:10.1525/bio.2009.59.5.9. S2CID 55591360. Archived from the original on 2011-08-07. Retrieved 2009-10-27.