Related Research Articles

Protein primary structure is the linear sequence of amino acids in a peptide or protein. By convention, the primary structure of a protein is reported starting from the amino-terminal (N) end to the carboxyl-terminal (C) end. Protein biosynthesis is most commonly performed by ribosomes in cells. Peptides can also be synthesized in the laboratory. Protein primary structures can be directly sequenced, or inferred from DNA sequences.

Lipoic acid (LA), also known as α-lipoic acid, alpha-lipoic acid (ALA) and thioctic acid, is an organosulfur compound derived from caprylic acid. ALA is made in animals normally, and is essential for aerobic metabolism. It is also manufactured and is available as a dietary supplement in some countries where it is marketed as an antioxidant, and is available as a pharmaceutical drug in other countries.

Post-translational modification (PTM) refers to the covalent and generally enzymatic modification of proteins following protein biosynthesis. Proteins are synthesized by ribosomes translating mRNA into polypeptide chains, which may then undergo PTM to form the mature protein product. PTMs are important components in cell signaling, as for example when prohormones are converted to hormones.
A prosthetic group is the non-amino acid component that is part of the structure of the heteroproteins or conjugated proteins, being covalently linked to the apoprotein.

Pyridoxal phosphate (PLP, pyridoxal 5'-phosphate, P5P), the active form of vitamin B6, is a coenzyme in a variety of enzymatic reactions. The International Union of Biochemistry and Molecular Biology has catalogued more than 140 PLP-dependent activities, corresponding to ~4% of all classified activities. The versatility of PLP arises from its ability to covalently bind the substrate, and then to act as an electrophilic catalyst, thereby stabilizing different types of carbanionic reaction intermediates.
In biochemistry, biotinylation is the process of covalently attaching biotin to a protein, nucleic acid or other molecule. Biotinylation is rapid, specific and is unlikely to disturb the natural function of the molecule due to the small size of biotin. Biotin binds to streptavidin and avidin with an extremely high affinity, fast on-rate, and high specificity, and these interactions are exploited in many areas of biotechnology to isolate biotinylated molecules of interest. Biotin-binding to streptavidin and avidin is resistant to extremes of heat, pH and proteolysis, making capture of biotinylated molecules possible in a wide variety of environments. Also, multiple biotin molecules can be conjugated to a protein of interest, which allows binding of multiple streptavidin, avidin or neutravidin protein molecules and increases the sensitivity of detection of the protein of interest. There is a large number of biotinylation reagents available that exploit the wide range of possible labelling methods. Due to the strong affinity between biotin and streptavidin, the purification of biotinylated proteins has been a widely used approach to identify protein-protein interactions and post-translational events such as ubiquitylation in molecular biology.

Pyruvate carboxylase (PC) encoded by the gene PC is an enzyme of the ligase class that catalyzes the physiologically irreversible carboxylation of pyruvate to form oxaloacetate (OAA).
Acetyl-CoA carboxylase (ACC) is a biotin-dependent enzyme that catalyzes the irreversible carboxylation of acetyl-CoA to produce malonyl-CoA through its two catalytic activities, biotin carboxylase (BC) and carboxyltransferase (CT). ACC is a multi-subunit enzyme in most prokaryotes and in the chloroplasts of most plants and algae, whereas it is a large, multi-domain enzyme in the cytoplasm of most eukaryotes. The most important function of ACC is to provide the malonyl-CoA substrate for the biosynthesis of fatty acids. The activity of ACC can be controlled at the transcriptional level as well as by small molecule modulators and covalent modification. The human genome contains the genes for two different ACCs—ACACA and ACACB.

A catalytic triad is a set of three coordinated amino acids that can be found in the active site of some enzymes. Catalytic triads are most commonly found in hydrolase and transferase enzymes. An Acid-Base-Nucleophile triad is a common motif for generating a nucleophilic residue for covalent catalysis. The residues form a charge-relay network to polarise and activate the nucleophile, which attacks the substrate, forming a covalent intermediate which is then hydrolysed to release the product and regenerate free enzyme. The nucleophile is most commonly a serine or cysteine amino acid, but occasionally threonine or even selenocysteine. The 3D structure of the enzyme brings together the triad residues in a precise orientation, even though they may be far apart in the sequence.
The branched-chain α-ketoacid dehydrogenase complex is a multi-subunit complex of enzymes that is found on the mitochondrial inner membrane. This enzyme complex catalyzes the oxidative decarboxylation of branched, short-chain alpha-ketoacids. BCKDC is a member of the mitochondrial α-ketoacid dehydrogenase complex family comprising pyruvate dehydrogenase and alpha-ketoglutarate dehydrogenase, key enzymes that function in the Krebs cycle.
Enzyme catalysis is the increase in the rate of a process by a biological molecule, an "enzyme". Most enzymes are proteins, and most such processes are chemical reactions. Within the enzyme, generally catalysis occurs at a localized site, called the active site.

In enzymology, an alanine racemase is an enzyme that catalyzes the chemical reaction

Cystathionine beta-lyase, also commonly referred to as CBL or β-cystathionase, is an enzyme that primarily catalyzes the following α,β-elimination reaction
Lipoyl synthase is an enzyme that belongs to the radical SAM (S-adenosylmethionine) family. Within the radical SAM superfamily, lipoyl synthase is in a sub-family of enzymes that catalyze sulfur insertion reactions. Enzymes in this family contain two 4Fe-4S clusters, from which they obtain the sulfur groups that will be transferred onto the corresponding substrates. This particular enzyme participates in lipoic acid metabolism, so it transfers two sulfurs from its 4Fe-4S cluster onto the protein N6-(octanoyl)lysine through radical generation. This enzyme is usually localized to the mitochondria. Two organisms that have been extensively studied with regards to this enzyme are Escherichia coli andMycobacterium tuberculosis. It is also found in other organisms, such as yeast and plants.

Acid-Induced Arginine Decarboxylase (AdiA), also commonly referred to as arginine decarboxylase, is an enzyme responsible for catalyzing the conversion of L-arginine into agmatine and carbon dioxide. The process consumes a proton in the decarboxylation and employs a pyridoxal-5'-phosphate (PLP) cofactor, similar to other enzymes involved in amino acid metabolism, such as ornithine decarboxylase and glutamine decarboxylase. It is found in bacteria and virus, though most research has so far focused on forms of the enzyme in bacteria. During the AdiA catalyzed decarboxylation of arginine, the necessary proton is consumed from the cell cytoplasm which helps to prevent the over-accumulation of protons inside the cell and serves to increase the intracellular pH. Arginine decarboxylase is part of an enzymatic system in Escherichia coli, Salmonella Typhimurium, and methane-producing bacteria Methanococcus jannaschii that makes these organisms acid resistant and allows them to survive under highly acidic medium.

In enzymology, diaminopimelate decarboxylase, also known as diaminopimelic acid decarboxylase, DAPDC, meso-diaminopimelate decarboxylase, DAP-decarboxylase, and meso-2,6-diaminoheptanedioate carboxy-lyase, is an enzyme that catalyzes the cleavage of carbon-carbon bonds in meso 2,6 diaminoheptanedioate to produce CO2 and L-lysine, the essential amino acid. It employs the cofactor pyridoxal phosphate, also known as PLP, which participates in numerous enzymatic transamination, decarboxylation and deamination reactions.
In enzymology, a lipoyl(octanoyl) transferase (EC 2.3.1.181) is an enzyme that catalyzes the chemical reaction

In enzymology, a serine C-palmitoyltransferase (EC 2.3.1.50) is an enzyme that catalyzes the chemical reaction:

In molecular biology, the Cofactor transferase family is a family of protein domains that includes biotin protein ligases, lipoate-protein ligases A, octanoyl-(acyl carrier protein):protein N-octanoyltransferases, and lipoyl-protein:protein N-lipoyltransferases. The metabolism of the cofactors Biotin and lipoic acid share this family. They also share the target modification domain, and the sulfur insertion enzyme.
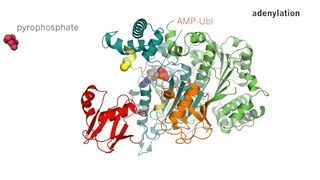
Lipoate–protein ligase (EC 2.7.7.63, LplA, lipoate protein ligase, lipoate–protein ligase A, LPL, LPL-B) is an enzyme with systematic name ATP:lipoate adenylyltransferase. This enzyme catalyses the following chemical reaction
References
- ↑ Kumar GK, Shenoy BC, Wood HG, Samols D, Xie Y, Park VL, Beegen H (1992). "The importance of methionine residues for the catalysis of the biotin enzyme, transcarboxylase. Analysis by site-directed mutagenesis". J. Biol. Chem. 267 (26): 18407–18412. doi: 10.1016/S0021-9258(19)36977-7 . PMID 1526981.
- ↑ Guest JR, Russell GC (1991). "Sequence similarities within the family of dihydrolipoamide acyltransferases and discovery of a previously unidentified fungal enzyme". Biochim. Biophys. Acta. 1076 (2): 225–232. doi:10.1016/0167-4838(91)90271-z. PMID 1825611.