Related Research Articles
Chromatin is a complex of DNA and protein found in eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important roles in reinforcing the DNA during cell division, preventing DNA damage, and regulating gene expression and DNA replication. During mitosis and meiosis, chromatin facilitates proper segregation of the chromosomes in anaphase; the characteristic shapes of chromosomes visible during this stage are the result of DNA being coiled into highly condensed chromatin.

In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn are wrapped into 30-nanometer fibers that form tightly packed chromatin. Histones prevent DNA from becoming tangled and protect it from DNA damage. In addition, histones play important roles in gene regulation and DNA replication. Without histones, unwound DNA in chromosomes would be very long. For example, each human cell has about 1.8 meters of DNA if completely stretched out; however, when wound about histones, this length is reduced to about 90 micrometers (0.09 mm) of 30 nm diameter chromatin fibers.
In molecular biology and genetics, transcriptional regulation is the means by which a cell regulates the conversion of DNA to RNA (transcription), thereby orchestrating gene activity. A single gene can be regulated in a range of ways, from altering the number of copies of RNA that are transcribed, to the temporal control of when the gene is transcribed. This control allows the cell or organism to respond to a variety of intra- and extracellular signals and thus mount a response. Some examples of this include producing the mRNA that encode enzymes to adapt to a change in a food source, producing the gene products involved in cell cycle specific activities, and producing the gene products responsible for cellular differentiation in multicellular eukaryotes, as studied in evolutionary developmental biology.

S phase (Synthesis Phase) is the phase of the cell cycle in which DNA is replicated, occurring between G1 phase and G2 phase. Since accurate duplication of the genome is critical to successful cell division, the processes that occur during S-phase are tightly regulated and widely conserved.

Charles David Allis was an American molecular biologist, and the Joy and Jack Fishman Professor at the Rockefeller University. He was also the Head of the Laboratory of Chromatin Biology and Epigenetics, and a professor at the Tri-Institutional MD–PhD Program.
The histone code is a hypothesis that the transcription of genetic information encoded in DNA is in part regulated by chemical modifications to histone proteins, primarily on their unstructured ends. Together with similar modifications such as DNA methylation it is part of the epigenetic code. Histones associate with DNA to form nucleosomes, which themselves bundle to form chromatin fibers, which in turn make up the more familiar chromosome. Histones are globular proteins with a flexible N-terminus that protrudes from the nucleosome. Many of the histone tail modifications correlate very well to chromatin structure and both histone modification state and chromatin structure correlate well to gene expression levels. The critical concept of the histone code hypothesis is that the histone modifications serve to recruit other proteins by specific recognition of the modified histone via protein domains specialized for such purposes, rather than through simply stabilizing or destabilizing the interaction between histone and the underlying DNA. These recruited proteins then act to alter chromatin structure actively or to promote transcription. For details of gene expression regulation by histone modifications see table below.

Eukaryotic transcription is the elaborate process that eukaryotic cells use to copy genetic information stored in DNA into units of transportable complementary RNA replica. Gene transcription occurs in both eukaryotic and prokaryotic cells. Unlike prokaryotic RNA polymerase that initiates the transcription of all different types of RNA, RNA polymerase in eukaryotes comes in three variations, each translating a different type of gene. A eukaryotic cell has a nucleus that separates the processes of transcription and translation. Eukaryotic transcription occurs within the nucleus where DNA is packaged into nucleosomes and higher order chromatin structures. The complexity of the eukaryotic genome necessitates a great variety and complexity of gene expression control.
Chromatin remodeling is the dynamic modification of chromatin architecture to allow access of condensed genomic DNA to the regulatory transcription machinery proteins, and thereby control gene expression. Such remodeling is principally carried out by 1) covalent histone modifications by specific enzymes, e.g., histone acetyltransferases (HATs), deacetylases, methyltransferases, and kinases, and 2) ATP-dependent chromatin remodeling complexes which either move, eject or restructure nucleosomes. Besides actively regulating gene expression, dynamic remodeling of chromatin imparts an epigenetic regulatory role in several key biological processes, egg cells DNA replication and repair; apoptosis; chromosome segregation as well as development and pluripotency. Aberrations in chromatin remodeling proteins are found to be associated with human diseases, including cancer. Targeting chromatin remodeling pathways is currently evolving as a major therapeutic strategy in the treatment of several cancers.

Histone H3.2 is a protein that in humans is encoded by the HIST2H3C gene.

Bromodomain-containing protein 4 is a protein that in humans is encoded by the BRD4 gene.
RNA polymerase II holoenzyme is a form of eukaryotic RNA polymerase II that is recruited to the promoters of protein-coding genes in living cells. It consists of RNA polymerase II, a subset of general transcription factors, and regulatory proteins known as SRB proteins.
Epigenomics is the study of the complete set of epigenetic modifications on the genetic material of a cell, known as the epigenome. The field is analogous to genomics and proteomics, which are the study of the genome and proteome of a cell. Epigenetic modifications are reversible modifications on a cell's DNA or histones that affect gene expression without altering the DNA sequence. Epigenomic maintenance is a continuous process and plays an important role in stability of eukaryotic genomes by taking part in crucial biological mechanisms like DNA repair. Plant flavones are said to be inhibiting epigenomic marks that cause cancers. Two of the most characterized epigenetic modifications are DNA methylation and histone modification. Epigenetic modifications play an important role in gene expression and regulation, and are involved in numerous cellular processes such as in differentiation/development and tumorigenesis. The study of epigenetics on a global level has been made possible only recently through the adaptation of genomic high-throughput assays.

Forkhead box protein A2 (FOXA2), also known as hepatocyte nuclear factor 3-beta (HNF-3B), is a transcription factor that plays an important role during development, in mature tissues and, when dysregulated or mutated, also in cancer.
The TBP-associated factors (TAF) are proteins that associate with the TATA-binding protein in transcription initiation. It is a part of the transcription initiation factor TFIID multimeric protein complex. It also makes up many other factors, including SL1. They mediate the formation of the transcription preinitiation complex, a step preceding transcription of DNA to RNA by RNA polymerase II.
Pioneer factors are transcription factors that can directly bind condensed chromatin. They can have positive and negative effects on transcription and are important in recruiting other transcription factors and histone modification enzymes as well as controlling DNA methylation. They were first discovered in 2002 as factors capable of binding to target sites on nucleosomal DNA in compacted chromatin and endowing competency for gene activity during hepatogenesis. Pioneer factors are involved in initiating cell differentiation and activation of cell-specific genes. This property is observed in histone fold-domain containing transcription factors and other transcription factors that use zinc finger(s) for DNA binding.
H3K4me3 is an epigenetic modification to the DNA packaging protein Histone H3 that indicates tri-methylation at the 4th lysine residue of the histone H3 protein and is often involved in the regulation of gene expression. The name denotes the addition of three methyl groups (trimethylation) to the lysine 4 on the histone H3 protein.
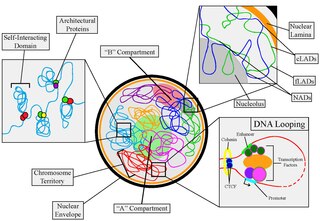
Nuclear organization refers to the spatial distribution of chromatin within a cell nucleus. There are many different levels and scales of nuclear organisation. Chromatin is a higher order structure of DNA.
H4K5ac is an epigenetic modification to the DNA packaging protein histone H4. It is a mark that indicates the acetylation at the 5th lysine residue of the histone H4 protein. H4K5 is the closest lysine residue to the N-terminal tail of histone H4. It is enriched at the transcription start site (TSS) and along gene bodies. Acetylation of histone H4K5 and H4K12ac is enriched at centromeres.
H3S10P is an epigenetic modification to the DNA packaging protein histone H3. It is a mark that indicates the phosphorylation the 10th serine residue of the histone H3 protein.
H3S28P is an epigenetic modification to the DNA packaging protein histone H3. It is a mark that indicates the phosphorylation the 28th serine residue of the histone H3 protein.
References
- ↑ Sarge, K. D.; Park-Sarge, O. K. (2005). "Gene bookmarking: Keeping the pages open". Trends in Biochemical Sciences. 30 (11): 605–10. doi:10.1016/j.tibs.2005.09.004. PMID 16188444.
- ↑ Martínez-Balbás, M. A.; Dey, A; Rabindran, S. K.; Ozato, K; Wu, C (1995). "Displacement of sequence-specific transcription factors from mitotic chromatin". Cell. 83 (1): 29–38. doi: 10.1016/0092-8674(95)90231-7 . PMID 7553870.
- ↑ John, S; Workman, J. L. (1998). "Bookmarking genes for activation in condensed mitotic chromosomes". BioEssays. 20 (4): 275–9. doi:10.1002/(SICI)1521-1878(199804)20:4<275::AID-BIES1>3.0.CO;2-P. PMID 9619097.
- ↑ Hsiung, C. C.; Morrissey, C. S.; Udugama, M; Frank, C. L.; Keller, C. A.; Baek, S; Giardine, B; Crawford, G. E.; Sung, M. H.; Hardison, R. C.; Blobel, G. A. (2015). "Genome accessibility is widely preserved and locally modulated during mitosis". Genome Research. 25 (2): 213–25. doi:10.1101/gr.180646.114. PMC 4315295 . PMID 25373146.
- ↑ Wang, F; Higgins, J. M. (2013). "Histone modifications and mitosis: Countermarks, landmarks, and bookmarks". Trends in Cell Biology. 23 (4): 175–84. doi:10.1016/j.tcb.2012.11.005. PMID 23246430.
- ↑ Kouskouti, A; Talianidis, I (2005). "Histone modifications defining active genes persist after transcriptional and mitotic inactivation". The EMBO Journal. 24 (2): 347–57. doi:10.1038/sj.emboj.7600516. PMC 545808 . PMID 15616580.
- ↑ Chow, C. M.; Georgiou, A; Szutorisz, H; Maia e Silva, A; Pombo, A; Barahona, I; Dargelos, E; Canzonetta, C; Dillon, N (2005). "Variant histone H3.3 marks promoters of transcriptionally active genes during mammalian cell division". EMBO Reports. 6 (4): 354–60. doi:10.1038/sj.embor.7400366. PMC 1299280 . PMID 15776021.
- ↑ Dey, A; Ellenberg, J; Farina, A; Coleman, A. E.; Maruyama, T; Sciortino, S; Lippincott-Schwartz, J; Ozato, K (2000). "A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G(2)-to-M transition". Molecular and Cellular Biology. 20 (17): 6537–49. doi:10.1128/mcb.20.17.6537-6549.2000. PMC 86127 . PMID 10938129.
- ↑ Dey, A; Nishiyama, A; Karpova, T; McNally, J; Ozato, K (2009). "Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription". Molecular Biology of the Cell. 20 (23): 4899–909. doi:10.1091/mbc.E09-05-0380. PMC 2785733 . PMID 19812244.
- ↑ Kadauke, S; Udugama, M. I.; Pawlicki, J. M.; Achtman, J. C.; Jain, D. P.; Cheng, Y; Hardison, R. C.; Blobel, G. A. (2012). "Tissue-specific mitotic bookmarking by hematopoietic transcription factor GATA1". Cell. 150 (4): 725–37. doi:10.1016/j.cell.2012.06.038. PMC 3425057 . PMID 22901805.
- ↑ Xing, H; Wilkerson, D. C.; Mayhew, C. N.; Lubert, E. J.; Skaggs, H. S.; Goodson, M. L.; Hong, Y; Park-Sarge, O. K.; Sarge, K. D. (2005). "Mechanism of hsp70i gene bookmarking". Science. 307 (5708): 421–3. Bibcode:2005Sci...307..421X. doi:10.1126/science.1106478. PMID 15662014.
- ↑ Christova, R; Oelgeschläger, T (2002). "Association of human TFIID-promoter complexes with silenced mitotic chromatin in vivo". Nature Cell Biology. 4 (1): 79–82. doi:10.1038/ncb733. PMID 11744923.
- ↑ Festuccia, N; Dubois, A; Vandormael-Pournin, S; Gallego Tejeda, E; Mouren, A; Bessonnard, S; Mueller, F; Proux, C; Cohen-Tannoudji, M; Navarro, P (November 2016). "Mitotic binding of Esrrb marks key regulatory regions of the pluripotency network" (PDF). Nature Cell Biology. 18 (11): 1139–1148. doi:10.1038/ncb3418. PMID 27723719.