
Bioinformatics is an interdisciplinary field that develops methods and software tools for understanding biological data, in particular when the data sets are large and complex. As an interdisciplinary field of science, bioinformatics combines biology, computer science, information engineering, mathematics and statistics to analyze and interpret the biological data. Bioinformatics has been used for in silico analyses of biological queries using mathematical and statistical techniques.

Metabolism is the set of life-sustaining chemical reactions in organisms. The three main purposes of metabolism are: the conversion of food to energy to run cellular processes; the conversion of food/fuel to building blocks for proteins, lipids, nucleic acids, and some carbohydrates; and the elimination of nitrogenous wastes. These enzyme-catalyzed reactions allow organisms to grow and reproduce, maintain their structures, and respond to their environments..
Computational biology involves the development and application of data-analytical and theoretical methods, mathematical modeling and computational simulation techniques to the study of biological, ecological, behavioral, and social systems. The field is broadly defined and includes foundations in biology, applied mathematics, statistics, biochemistry, chemistry, biophysics, molecular biology, genetics, genomics, computer science, and evolution.

A generegulatory network (GRN) is a collection of molecular regulators that interact with each other and with other substances in the cell to govern the gene expression levels of mRNA and proteins. These play a central role in morphogenesis, the creation of body structures, which in turn is central to evolutionary developmental biology (evo-devo).
The metabolic theory of ecology (MTE) is an extension of Kleiber's law and posits that the metabolic rate of organisms is the fundamental biological rate that governs most observed patterns in ecology. MTE is part of a larger set of theory known as metabolic scaling theory that attempts to provide a unified theory for the importance of metabolism in driving pattern and process in biology from the level of cells all the way to the biosphere.
Modelling biological systems is a significant task of systems biology and mathematical biology. Computational systems biology aims to develop and use efficient algorithms, data structures, visualization and communication tools with the goal of computer modelling of biological systems. It involves the use of computer simulations of biological systems, including cellular subsystems, to both analyze and visualize the complex connections of these cellular processes.

Metabolic network reconstruction and simulation allows for an in-depth insight into the molecular mechanisms of a particular organism. In particular, these models correlate the genome with molecular physiology. A reconstruction breaks down metabolic pathways into their respective reactions and enzymes, and analyzes them within the perspective of the entire network. In simplified terms, a reconstruction collects all of the relevant metabolic information of an organism and compiles it in a mathematical model. Validation and analysis of reconstructions can allow identification of key features of metabolism such as growth yield, resource distribution, network robustness, and gene essentiality. This knowledge can then be applied to create novel biotechnology.

KEGG is a collection of databases dealing with genomes, biological pathways, diseases, drugs, and chemical substances. KEGG is utilized for bioinformatics research and education, including data analysis in genomics, metagenomics, metabolomics and other omics studies, modeling and simulation in systems biology, and translational research in drug development.
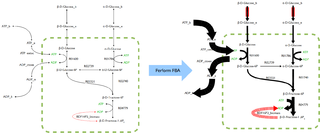
Flux balance analysis (FBA) is a mathematical method for simulating metabolism in genome-scale reconstructions of metabolic networks. In comparison to traditional methods of modeling, FBA is less intensive in terms of the input data required for constructing the model. Simulations performed using FBA are computationally inexpensive and can calculate steady-state metabolic fluxes for large models in a few seconds on modern personal computers.
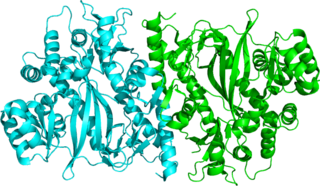
Glutathione synthetase (GSS) is the second enzyme in the glutathione (GSH) biosynthesis pathway. It catalyses the condensation of gamma-glutamylcysteine and glycine, to form glutathione. Glutathione synthetase is also a potent antioxidant. It is found in many species including bacteria, yeast, mammals, and plants.
Fluxomics describes the various approaches that seek to determine the rates of metabolic reactions within a biological entity. While metabolomics can provide instantaneous information on the metabolites in a biological sample, metabolism is a dynamic process. The significance of fluxomics is that metabolic fluxes determine the cellular phenotype. It has the added advantage of being based on the metabolome which has fewer components than the genome or proteome.

Gustavo Caetano-Anollés is Professor of Bioinformatics in the Department of Crop Sciences, University of Illinois at Urbana-Champaign. He is an expert in the field of evolutionary and comparative genomics.
The Molecular Ancestry Network (MANET) database is a bioinformatics database that maps evolutionary relationships of protein architectures directly onto biological networks. It was originally developed by Hee Shin Kim, Jay E. Mittenthal and Gustavo Caetano-Anolles in the Department of Crop Sciences of the University of Illinois at Urbana-Champaign.
Biology data visualization is a branch of bioinformatics concerned with the application of computer graphics, scientific visualization, and information visualization to different areas of the life sciences. This includes visualization of sequences, genomes, alignments, phylogenies, macromolecular structures, systems biology, microscopy, and magnetic resonance imaging data. Software tools used for visualizing biological data range from simple, standalone programs to complex, integrated systems.
Within biological systems, degeneracy occurs when structurally dissimilar components/modules/pathways can perform similar functions under certain conditions, but perform distinct functions in other conditions. Degeneracy is thus a relational property that requires comparing the behavior of two or more components. In particular, if degeneracy is present in a pair of components, then there will exist conditions where the pair will appear functionally redundant but other conditions where they will appear functionally distinct.

Robustness of a biological system is the persistence of a certain characteristic or trait in a system under perturbations or conditions of uncertainty. Robustness in development is known as canalization. According to the kind of perturbation involved, robustness can be classified as mutational, environmental, recombinational, or behavioral robustness etc. Robustness is achieved through the combination of many genetic and molecular mechanisms and can evolve by either direct or indirect selection. Several model systems have been developed to experimentally study robustness and its evolutionary consequences.

Systrip is a visual environment for the analysis of time-series data in the context of biological networks. Systrip gathers bioinformatics and graph theoretical algorithms that can be assembled in different ways to help biologists in their visual mining process. It had been used to analyze various real biological data.

Stefan Schuster is a German biophysicist. He is professor for bioinformatics at the University of Jena.
Pharmaceutical Bioinformatics is a research field related to bioinformatics but with the focus on studying biological and chemical processes in the pharmaceutical area; to understand how xenobiotics interact with the human body and the drug discovery process.
Machine learning, a subfield of computer science involving the development of algorithms that learn how to make predictions based on data, has a number of emerging applications in the field of bioinformatics. Bioinformatics deals with computational and mathematical approaches for understanding and processing biological data.











