DnaB helicase is an enzyme in bacteria which opens the replication fork during DNA replication. Although the mechanism by which DnaB both couples ATP hydrolysis to translocation along DNA and denatures the duplex is unknown, a change in the quaternary structure of the protein involving dimerisation of the N-terminal domain has been observed and may occur during the enzymatic cycle. Initially when DnaB binds to dnaA, it is associated with dnaC, a negative regulator. After DnaC dissociates, DnaB binds dnaG.
In biology and biochemistry, protease inhibitors, or antiproteases, are molecules that inhibit the function of proteases. Many naturally occurring protease inhibitors are proteins.

A leucine-rich repeat (LRR) is a protein structural motif that forms an α/β horseshoe fold. It is composed of repeating 20–30 amino acid stretches that are unusually rich in the hydrophobic amino acid leucine. These tandem repeats commonly fold together to form a solenoid protein domain, termed leucine-rich repeat domain. Typically, each repeat unit has beta strand-turn-alpha helix structure, and the assembled domain, composed of many such repeats, has a horseshoe shape with an interior parallel beta sheet and an exterior array of helices. One face of the beta sheet and one side of the helix array are exposed to solvent and are therefore dominated by hydrophilic residues. The region between the helices and sheets is the protein's hydrophobic core and is tightly sterically packed with leucine residues.

Carbamoyl phosphate synthetase catalyzes the ATP-dependent synthesis of carbamoyl phosphate from glutamine or ammonia and bicarbonate. This ATP-grasp enzyme catalyzes the reaction of ATP and bicarbonate to produce carboxy phosphate and ADP. Carboxy phosphate reacts with ammonia to give carbamic acid. In turn, carbamic acid reacts with a second ATP to give carbamoyl phosphate plus ADP.
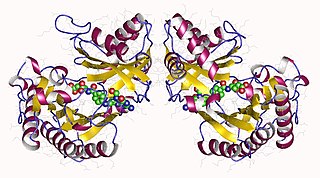
Glutamate formimidoyltransferase is a methyltransferase enzyme which uses tetrahydrofolate as part of histidine catabolism. It catalyses two reactions:

In enzymology, a protein-glutamate O-methyltransferase is an enzyme that catalyzes the chemical reaction

In enzymology, an ATP phosphoribosyltransferase is an enzyme that catalyzes the chemical reaction

The Rel homology domain (RHD) is a protein domain found in a family of eukaryotic transcription factors, including both NF-κB and NFAT, among others. Some of these transcription factors appear to form multi-protein DNA-bound complexes. Phosphorylation of the RHD appears to play a role in the regulation of some of these transcription factors, acting to modulate the expression of their target genes.

In molecular biology, phosphotyrosine-binding domains are protein domains which bind to phosphotyrosine.

In molecular biology, chaperone DnaJ, also known as Hsp40, is a molecular chaperone protein. It is expressed in a wide variety of organisms from bacteria to humans.

In molecular biology, the AMMECR1 protein is a protein encoded by the AMMECR1 gene on human chromosome Xq22.3.

In molecular biology, the ars operon is an operon found in several bacterial taxon. It is required for the detoxification of arsenate, arsenite, and antimonite. This system transports arsenite and antimonite out of the cell. The pump is composed of two polypeptides, the products of the arsA and arsB genes. This two-subunit enzyme produces resistance to arsenite and antimonite. Arsenate, however, must first be reduced to arsenite before it is extruded. A third gene, arsC, expands the substrate specificity to allow for arsenate pumping and resistance. ArsC is an approximately 150-residue arsenate reductase that uses reduced glutathione (GSH) to convert arsenate to arsenite with a redox active cysteine residue in the active site. ArsC forms an active quaternary complex with GSH, arsenate, and glutaredoxin 1 (Grx1). The three ligands must be present simultaneously for reduction to occur.
In molecular biology, autophagy related 3 (Atg3) is the E2 enzyme for the LC3 lipidation process. It is essential for autophagy. The super protein complex, the Atg16L complex, consists of multiple Atg12-Atg5 conjugates. Atg16L has an E3-like role in the LC3 lipidation reaction. The activated intermediate, LC3-Atg3 (E2), is recruited to the site where the lipidation takes place.
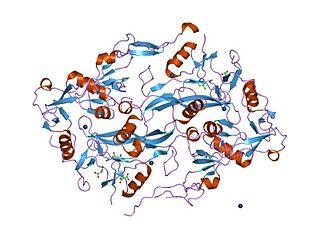
The Kazal domain is an evolutionary conserved protein domain usually indicative of serine protease inhibitors. However, kazal-like domains are also seen in the extracellular part of agrins, which are not known to be protease inhibitors.

In molecular biology, a carbohydrate-binding module (CBM) is a protein domain found in carbohydrate-active enzymes. The majority of these domains have carbohydrate-binding activity. Some of these domains are found on cellulosomal scaffoldin proteins. CBMs were previously known as cellulose-binding domains. CBMs are classified into numerous families, based on amino acid sequence similarity. There are currently 64 families of CBM in the CAZy database.
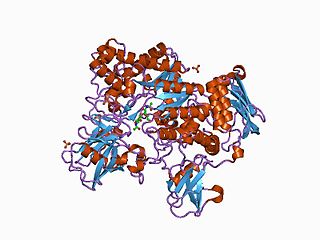
In molecular biology, the CHB HEX N-terminal domain represents the N-terminal domain in chitobiases and beta-hexosaminidases. Chitobiases degrade chitin, which forms the exoskeleton in insects and crustaceans, and which is one of the most abundant polysaccharides on earth. Beta-hexosaminidases are composed of either a HexA/HexB heterodimer or a HexB homodimer, and can hydrolyse diverse substrates, including GM(2)-gangliosides; mutations in this enzyme are associated with Tay–Sachs disease. HexB is structurally similar to chitobiase, consisting of a beta sandwich structure; this structure is similar to that found in the cellulose-binding domain of cellulase from Cellulomonas fimi. This domain may function as a carbohydrate binding module.

In molecular biology, the chitinase A N-terminal domain is found at the N-terminus of a number of bacterial chitinases and similar viral proteins. It is organised into a fibronectin III module domain-like fold, comprising only beta strands. Its function is not known, but it may be involved in interaction with the enzyme substrate, chitin. It is separated by a hinge region from the catalytic domain; this hinge region is probably mobile, allowing the N-terminal domain to have different relative positions in solution.
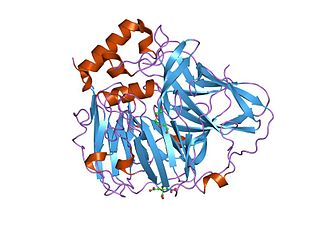
In molecular biology, multicopper oxidases are enzymes which oxidise their substrate by accepting electrons at a mononuclear copper centre and transferring them to a trinuclear copper centre; dioxygen binds to the trinuclear centre and, following the transfer of four electrons, is reduced to two molecules of water. There are three spectroscopically different copper centres found in multicopper oxidases: type 1, type 2 and type 3. Multicopper oxidases consist of 2, 3 or 6 of these homologous domains, which also share homology with the cupredoxins azurin and plastocyanin. Structurally, these domains consist of a cupredoxin-like fold, a beta-sandwich consisting of 7 strands in 2 beta-sheets, arranged in a Greek-key beta-barrel.

In molecular biology, D-stereospecific aminopeptidase (D-aminopeptidase) EC 3.4.11.19 is an enzyme which catalyses the release of an N-terminal D-amino acid from a peptide, Xaa-|-Yaa-, in which Xaa is preferably D-Ala, D-Ser or D-Thr. D-amino acid amides and methyl esters also are hydrolyzed, as is glycine amide.
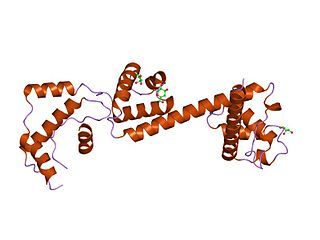
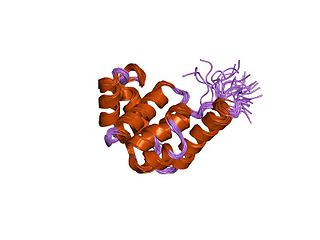
In molecular biology, the HAND domain is a protein domain which adopts a secondary structure consisting of four alpha helices, three of which form an L-like configuration. Helix H2 runs antiparallel to helices H3 and H4, packing closely against helix H4, whilst helix H1 reposes in the concave surface formed by these three helices and runs perpendicular to them. This domain confers DNA and nucleosome binding properties to the proteins in which it occurs. It is named the HAND domain because its 4-helical structure resembles an open hand.



















