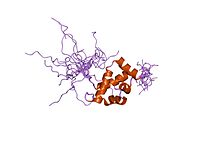
A homeobox is a DNA sequence, around 180 base pairs long, found within genes that are involved in the regulation of patterns of anatomical development (morphogenesis) in animals, fungi and plants. These genes encode homeodomain protein products that are transcription factors sharing a characteristic protein fold structure that binds DNA. The "homeo-" prefix in the words "homeobox" and "homeodomain" stems from the mutational phenotype known as "homeosis", which is frequently observed when these genes are mutated in animals. Homeosis is a term coined by William Bateson to describe the outright replacement of a discrete body part with another body part. Homeobox genes are not only found in animals, but have also been found in fungi, for example the unicellular yeasts, in plants, and numerous single cell eukaryotes.
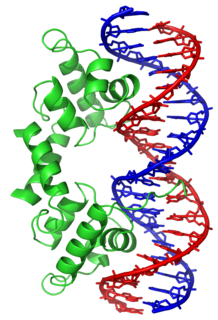
In proteins, the helix-turn-helix (HTH) is a major structural motif capable of binding DNA. Each monomer incorporates two α helices, joined by a short strand of amino acids, that bind to the major groove of DNA. The HTH motif occurs in many proteins that regulate gene expression. It should not be confused with the helix-loop-helix motif.
Hepatocyte nuclear factors (HNFs) are a group of phylogenetically unrelated transcription factors that regulate the transcription of a diverse group of genes into proteins. These proteins include blood clotting factors and in addition, enzymes and transporters involved with glucose, cholesterol, and fatty acid transport and metabolism.
The W box is a deoxyribonucleic acid (DNA) cis-regulatory element sequence, (T)TGAC(C/T), which is recognized by the family of WRKY transcription factors.

Apetala 2(AP2) is a gene and a member of a large family of transcription factors, the AP2/EREBP family. In Arabidopsis thaliana AP2 plays a role in the ABC model of flower development. It was originally thought that this family of proteins was plant-specific; however, recent studies have shown that apicomplexans, including the causative agent of malaria, Plasmodium falciparum encode a related set of transcription factors, called the ApiAP2 family.

Transcription factor 3, also known as TCF3, is a protein that in humans is encoded by the TCF3 gene. TCF3 has been shown to directly enhance Hes1 expression.

POU domain, class 2, transcription factor 1 is a protein that in humans is encoded by the POU2F1 gene.

Interleukin enhancer-binding factor 3 is a protein that in humans is encoded by the ILF3 gene.

Pre-B-cell leukemia transcription factor 1 is a protein that in humans is encoded by the PBX1 gene.

Y box binding protein 1 also known as Y-box transcription factor or nuclease-sensitive element-binding protein 1 is a protein that in humans is encoded by the YBX1 gene.
POU is a family of proteins that have well-conserved homeodomains.

Cux1 is a homeodomain protein that in humans is encoded by the CUX1 gene.

Homeobox protein Meis1 is a protein that in humans is encoded by the MEIS1 gene.

LIM domain-binding protein 1 is a protein that in humans is encoded by the LDB1 gene.

LIM/homeobox protein Lhx3 is a protein that in humans is encoded by the LHX3 gene.

Interleukin enhancer-binding factor 2 is a protein that in humans is encoded by the ILF2 gene.
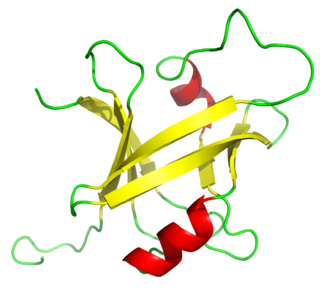
The B3 DNA binding domain (DBD) is a highly conserved domain found exclusively in transcription factors, from higher plants combined with other domains. It consists of 100-120 residues, includes seven beta strands and two alpha helices that form a DNA-binding pseudobarrel protein fold ; it interacts with the major groove of DNA.
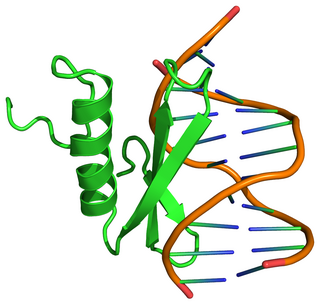
Ethylene-responsive element binding protein(EREBP) is a homeobox gene from Arabidopsis thaliana and other plants which encodes a transcription factor. EREBP is responsible in part for mediating the response in plants to the plant hormone ethylene.

The SQUAMOSA promoter binding protein-like family of transcription factors are defined by a plant-specific DNA-binding domain. The founding member of the family was identified based on its specific in vitro binding to the promoter of the snapdragon SQUAMOSA gene. SBP proteins are thought to be transcriptional activators.

The WRKY domain is found in the WRKY transcription factor family, a class of transcription factors. The WRKY domain is found almost exclusively in plants although WRKY genes appear present in some diplomonads, social amoebae and other amoebozoa, and fungi incertae sedis. They appear absent in other non-plant species. WRKY transcription factors have been a significant area of plant research for the past 20 years. The WRKY DNA-binding domain recognizes the W-box (T)TGAC(C/T) cis-regulatory element.
The public domain consists of all the creative works to which no exclusive intellectual property rights apply. Those rights may have expired, been forfeited, expressly waived, or may be inapplicable.

Pfam is a database of protein families that includes their annotations and multiple sequence alignments generated using hidden Markov models. The most recent version, Pfam 31.0, was released in March 2017 and contains 16,712 families.
InterPro is a database of protein families, domains and functional sites in which identifiable features found in known proteins can be applied to new protein sequences in order to functionally characterise them.