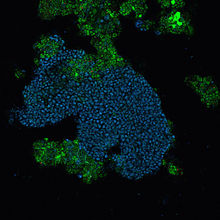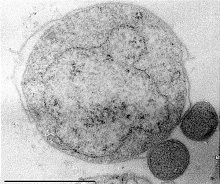
Nanoarchaeota are a phylum of the Archaea. This phylum currently has only one representative, Nanoarchaeum equitans.
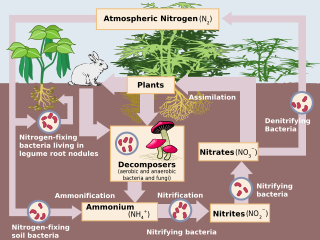
Denitrification is a microbially facilitated process where nitrate (NO3−) is reduced and ultimately produces molecular nitrogen (N2) through a series of intermediate gaseous nitrogen oxide products. Facultative anaerobic bacteria perform denitrification as a type of respiration that reduces oxidized forms of nitrogen in response to the oxidation of an electron donor such as organic matter. The preferred nitrogen electron acceptors in order of most to least thermodynamically favorable include nitrate (NO3−), nitrite (NO2−), nitric oxide (NO), nitrous oxide (N2O) finally resulting in the production of dinitrogen (N2) completing the nitrogen cycle. Denitrifying microbes require a very low oxygen concentration of less than 10%, as well as organic C for energy. Since denitrification can remove NO3−, reducing its leaching to groundwater, it can be strategically used to treat sewage or animal residues of high nitrogen content. Denitrification can leak N2O, which is an ozone-depleting substance and a greenhouse gas that can have a considerable influence on global warming.

Acidobacteria is a phylum of bacteria. Its members are physiologically diverse and ubiquitous, especially in soils, but are under-represented in culture.

Anammox, an abbreviation for anaerobic ammonium oxidation, is a globally important microbial process of the nitrogen cycle that takes place in many natural environments. The bacteria mediating this process were identified in 1999, and were a great surprise for the scientific community. In the anammox reaction, nitrite and ammonium ions are converted directly into diatomic nitrogen and water.
Biological augmentation is the addition of archaea or bacterial cultures required to speed up the rate of degradation of a contaminant. Organisms that originate from contaminated areas may already be able to break down waste, but perhaps inefficiently and slowly.

A phytase is any type of phosphatase enzyme that catalyzes the hydrolysis of phytic acid – an indigestible, organic form of phosphorus that is found in many plant tissues, especially in grains and oil seeds – and releases a usable form of inorganic phosphorus. While phytases have been found to occur in animals, plants, fungi and bacteria, phytases have been most commonly detected and characterized from fungi.
Enhanced biological phosphorus removal (EBPR) is a sewage treatment configuration applied to activated sludge systems for the removal of phosphate.

Polyphosphate-accumulating organisms (PAOs) are a group of bacteria that, under certain conditions, facilitate the removal of large amounts of phosphorus from wastewater in a process, called enhanced biological phosphorus removal (EBPR). PAOs accomplish this removal of phosphate by accumulating it within their cells as polyphosphate. PAOs are by no means the only bacteria that can accumulate polyphosphate within their cells and in fact, the production of polyphosphate is a widespread ability among bacteria. However, the PAOs have many characteristics that other organisms that accumulate polyphosphate do not have, that make them amenable to use in wastewater treatment. Specifically, this is the ability to consume simple carbon compounds without the presence of an external electron acceptor by generating energy from internally stored polyphosphate and glycogen. Most other bacteria cannot consume under these conditions and therefore PAOs gain a selective advantage within the mixed microbial community present in the activated sludge. Therefore, wastewater treatment plants that operate for enhanced biological phosphorus removal have an anaerobic tank prior to the other tanks to give PAOs preferential access to the simple carbon compounds in the wastewater that is influent to the plant.

Alphaproteobacteria is a class of bacteria in the phylum Proteobacteria. Its members are highly diverse and possess few commonalities, but nevertheless share a common ancestor. Like all Proteobacteria, its members are gram-negative and some of its intracellular parasitic members lack peptidoglycan and are consequently gram variable.
The Gemmatimonadetes are a phylum of bacteria established in 2003. The phylum contains two classes Gemmatimonadetes and Longimicrobia.
Comamonas testosteroni is a Gram-negative soil bacterium. Strain I2gfp has been used in bioaugmentation trials, in attempts to treat the industrial byproduct 3-chloroaniline.
In enzymology, a xanthan lyase is an enzyme that catalyzes the chemical reaction of cleaving the beta-D-mannosyl-beta-D-1,4-glucuronosyl bond on the polysaccharide xanthan. This enzyme belongs to the family of lyases, specifically those carbon-oxygen lyases acting on polysaccharides. Xanthan lyase was first identified and partially purified in 1987.

Class II bacteriocins are a class of small peptides that inhibit the growth of various bacteria.
Sakacins are bacteriocins produced by Lactobacillus sakei. They are often clustered with the other lactic acid bacteriocins. The best known sakacins are sakacin A, G, K, P, and Q. In particular, sakacin A and P have been well characterized.
Elusimicrobium minutum is an ultramicrobacterium and first accepted member to be cultured of a major bacterial lineage previously known only as candidate phylum Termite Gut 1 (TG1), which has accordingly been renamed phylum Elusimicrobia. It was isolated in the laboratory of Andreas Brune at the Max Planck Institute for Terrestrial Microbiology, from the scarab beetle. It is a mesophilic, obligately anaerobic ultramicrobacterium with a gram-negative cell envelope. Cells are typically rod shaped, but cultures are pleomorphic in all growth phases. The isolate grows heterotrophically on sugars and ferments D-galactose, D-glucose, D-fructose, D-glucosamine, and N-acetyl-D-glucosamine to acetate, ethanol, hydrogen, and alanine as major products but only if amino acids are present in the medium
The phylum Elusimicrobia, previously known as "Termite Group 1", has been shown to be widespread in different ecosystems like marine environment, sewage sludge, contaminated sites and soils, and toxic wastes. The high abundance of 'Elusimicrobia' representatives is only evidenced for the lineage of symbionts found in termites and ants.
Saccharibacteria, formerly known as TM7, is a major bacterial lineage. It was discovered through 16S rRNA sequencing.
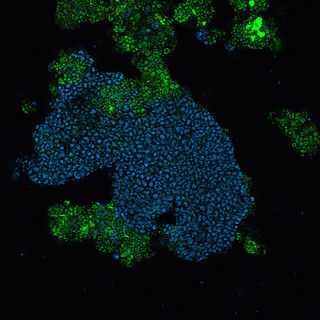
"Candidatus Accumulibacter" is an unclassified group of Betaproteobacteria that currently contains only a single member, "Candidatus Accumulibacter phosphatis". "Ca. A. phosphatis" is a common bacterial community member of wastewater treatment plants performing enhanced biological phosphorus removal and is a polyphosphate-accumulating organism. There are currently no cultured representatives, however due to the importance of "Ca. A. phosphatis" in the biotechnology industry there has been much research into the physiology of these bacteria.
Koribacter versatilis is a member of the acidobacteria phylum which itself is a newly devised phylum of bacteria, and is only distantly related to other organisms in the domain bacteria. Its closest phylogenetic relative is candidatus Solibacter usitatus, according to Michael Nerdahl. It contains 5,650,368 nucleotides, 4,777 proteins, and 55 RNA genes, and has a circular chromosome according to information found from GenBank. According to the Joint Genome Institute, “The bacterium is a gram-negative, highly capsulated, aerobic heterotroph that grows with a range of sugars, sugar polymers, and some organic acids.”
Modulibacteria is a bacterial phylum formerly known as KS3B3 or GN06. It is a candidate phylum, meaning there are no cultured representatives of this group. Members of the Modulibacteria phylum are known to cause fatal filament overgrowth (bulking) in high-rate industrial anaerobic wastewater treatment bioreactors.