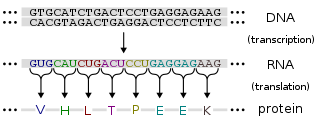
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product. These products are often proteins, but in non-protein coding genes such as transfer RNA (tRNA) or small nuclear RNA (snRNA) genes, the product is a functional RNA.

The lac repressor is a DNA-binding protein that inhibits the expression of genes coding for proteins involved in the metabolism of lactose in bacteria. These genes are repressed when lactose is not available to the cell, ensuring that the bacterium only invests energy in the production of machinery necessary for uptake and utilization of lactose when lactose is present. When lactose becomes available, it is converted into allolactose, which inhibits the lac repressor's DNA binding ability, thereby increasing gene expression.

DNA-binding proteins are proteins that have DNA-binding domains and thus have a specific or general affinity for single- or double-stranded DNA. Sequence-specific DNA-binding proteins generally interact with the major groove of B-DNA, because it exposes more functional groups that identify a base pair. However, there are some known minor groove DNA-binding ligands such as netropsin, distamycin, Hoechst 33258, pentamidine, DAPI and others.

Sterol regulatory element-binding proteins (SREBPs) are transcription factors that bind to the sterol regulatory element DNA sequence TCACNCCAC. Mammalian SREBPs are encoded by the genes SREBF1 and SREBF2. SREBPs belong to the basic-helix-loop-helix leucine zipper class of transcription factors. Unactivated SREBPs are attached to the nuclear envelope and endoplasmic reticulum membranes. In cells with low levels of sterols, SREBPs are cleaved to a water-soluble N-terminal domain that is translocated to the nucleus. These activated SREBPs then bind to specific sterol regulatory element DNA sequences, thus upregulating the synthesis of enzymes involved in sterol biosynthesis. Sterols in turn inhibit the cleavage of SREBPs and therefore synthesis of additional sterols is reduced through a negative feed back loop.
The Open Regulatory Annotation Database is designed to promote community-based curation of regulatory information. Specifically, the database contains information about regulatory regions, transcription factor binding sites, regulatory variants, and haplotypes.
Sterol regulatory element-binding transcription factor 1 (SREBF1) also known as sterol regulatory element-binding protein 1 (SREBP-1) is a protein that in humans is encoded by the SREBF1 gene.

Sterol regulatory element-binding protein 2 (SREBP-2) also known as sterol regulatory element binding transcription factor 2 (SREBF2) is a protein that in humans is encoded by the SREBF2 gene.
Membrane-bound transcription factor site-2 protease, or site-2 protease (S2P) for short, is an enzyme encoded by the MBTPS2 gene which liberates the N-terminal fragment of sterol regulatory element-binding protein (SREBP) transcription factors from membranes. S2P cleaves the transmembrane domain of SREPB, making it a member of the class of intramembrane proteases.
ChIP-sequencing, also known as ChIP-seq, is a method used to analyze protein interactions with DNA. ChIP-seq combines chromatin immunoprecipitation (ChIP) with massively parallel DNA sequencing to identify the binding sites of DNA-associated proteins. It can be used to map global binding sites precisely for any protein of interest. Previously, ChIP-on-chip was the most common technique utilized to study these protein–DNA relations.

The bacterial one-hybrid (B1H) system is a method for identifying the sequence-specific target site of a DNA-binding domain. In this system, a given transcription factor (TF) is expressed as a fusion to a subunit of RNA polymerase. In parallel, a library of randomized oligonucleotides representing potential TF target sequences are cloned into a separate vector containing the selectable genes HIS3 and URA3. If the DNA-binding domain (bait) binds a potential DNA target site (prey) in vivo, it will recruit RNA polymerase to the promoter and activate transcription of the reporter genes in that clone. The two reporter genes, HIS3 and URA3, allow for positive and negative selections, respectively. At the end of the process, positive clones are sequenced and examined with motif-finding tools in order to resolve the favoured DNA target sequence.
Cis-regulatory module (CRM) is a stretch of DNA, usually 100–1000 DNA base pairs in length, where a number of transcription factors can bind and regulate expression of nearby genes and regulate their transcription rates. They are labeled as cis because they are typically located on the same DNA strand as the genes they control as opposed to trans, which refers to effects on genes not located on the same strand or farther away, such as transcription factors. One cis-regulatory element can regulate several genes, and conversely, one gene can have several cis-regulatory modules.Cis-regulatory modules carry out their function by integrating the active transcription factors and the associated co-factors at a specific time and place in the cell where this information is read and an output is given.
DNA binding sites are a type of binding site found in DNA where other molecules may bind. DNA binding sites are distinct from other binding sites in that (1) they are part of a DNA sequence and (2) they are bound by DNA-binding proteins. DNA binding sites are often associated with specialized proteins known as transcription factors, and are thus linked to transcriptional regulation. The sum of DNA binding sites of a specific transcription factor is referred to as its cistrome. DNA binding sites also encompasses the targets of other proteins, like restriction enzymes, site-specific recombinases and methyltransferases.
RegulonDB is a database of the regulatory network of gene expression in Escherichia coli K-12. RegulonDB also models the organization of the genes in transcription units, operons and regulons. A total of 120 sRNAs with 231 total interactions which all together regulate 192 genes are also included. RegulonDB was founded in 1998 and also contributes data to the EcoCyc database.
YEASTRACT is a curated repository of more than 48000 regulatory associations between transcription factors (TF) and target genes in Saccharomyces cerevisiae, based on more than 1200 bibliographic references. It also includes the description of about 300 specific DNA binding sites for more than a hundred characterized TFs. Further information about each Yeast gene has been extracted from the Saccharomyces Genome Database (SGD). For each gene the associated Gene Ontology (GO) terms and their hierarchy in GO was obtained from the GO consortium. Currently, YEASTRACT maintains more than 7100 terms from GO. The nucleotide sequences of the promoter and coding regions for Yeast genes were obtained from Regulatory Sequence Analysis Tools (RSAT). All the information in YEASTRACT is updated regularly to match the latest data from SGD, GO consortium, RSA Tools and recent literature on yeast regulatory networks.
TRANSFAC is a manually curated database of eukaryotic transcription factors, their genomic binding sites and DNA binding profiles. The contents of the database can be used to predict potential transcription factor binding sites.
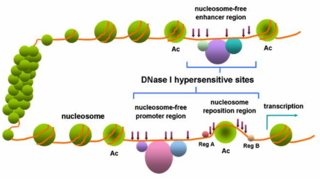
In genetics, DNase I hypersensitive sites (DHSs) are regions of chromatin that are sensitive to cleavage by the DNase I enzyme. In these specific regions of the genome, chromatin has lost its condensed structure, exposing the DNA and making it accessible. This raises the availability of DNA to degradation by enzymes, such as DNase I. These accessible chromatin zones are functionally related to transcriptional activity, since this remodeled state is necessary for the binding of proteins such as transcription factors.
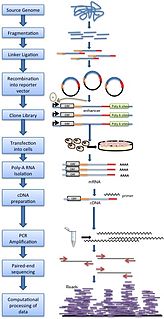
STARR-seq is a novel method to assay enhancer activity for millions of candidates from arbitrary sources of DNA. It is used to identify the sequences that act as transcriptional enhancers in a direct, quantitative, and genome-wide manner.
Identification of genomic regulatory elements is essential for understanding the dynamics of developmental, physiological and pathological processes. Recent advances in chromatin immunoprecipitation followed by sequencing (ChIP-seq) have provided powerful ways to identify genome-wide profiling of DNA-binding proteins and histone modifications. The application of ChIP-seq methods has reliably discovered transcription factor binding sites and histone modification sites.
JASPAR is an open access and widely used database of manually curated, non-redundant transcription factor (TF) binding profiles stored as position frequency matrices (PFM) and transcription factor flexible models (TFFM) for TFs from species in six taxonomic groups. From the supplied PFMs, users may generate position-specific weight matrices (PWM). The JASPAR database was introduced in 2004. There were five major updates and new releases in 2006, 2008, 2010, 2014, 2016 and 2018, which is the latest release of JASPAR.