Related Research Articles
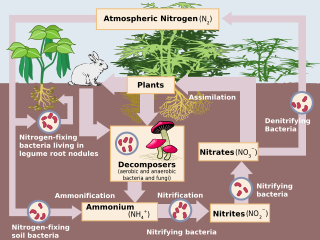
Nitrification is the biological oxidation of ammonia to nitrate via the intermediary nitrite. Nitrification is an important step in the nitrogen cycle in soil. The process of complete nitrification may occur through separate organisms or entirely within one organism, as in comammox bacteria. The transformation of ammonia to nitrite is usually the rate limiting step of nitrification. Nitrification is an aerobic process performed by small groups of autotrophic bacteria and archaea.

Bioremediation broadly refers to any process wherein a biological system, living or dead, is employed for removing environmental pollutants from air, water, soil, flue gasses, industrial effluents etc., in natural or artificial settings. The natural ability of organisms to adsorb, accumulate, and degrade common and emerging pollutants has attracted the use of biological resources in treatment of contaminated environment. In comparison to conventional physicochemical treatment methods bioremediation may offer advantages as it aims to be sustainable, eco-friendly, cheap, and scalable.
Methanotrophs are prokaryotes that metabolize methane as their source of carbon and chemical energy. They are bacteria or archaea, can grow aerobically or anaerobically, and require single-carbon compounds to survive.

Sulfate-reducing microorganisms (SRM) or sulfate-reducing prokaryotes (SRP) are a group composed of sulfate-reducing bacteria (SRB) and sulfate-reducing archaea (SRA), both of which can perform anaerobic respiration utilizing sulfate (SO2−
4) as terminal electron acceptor, reducing it to hydrogen sulfide (H2S). Therefore, these sulfidogenic microorganisms "breathe" sulfate rather than molecular oxygen (O2), which is the terminal electron acceptor reduced to water (H2O) in aerobic respiration.
Methylotrophs are a diverse group of microorganisms that can use reduced one-carbon compounds, such as methanol or methane, as the carbon source for their growth; and multi-carbon compounds that contain no carbon-carbon bonds, such as dimethyl ether and dimethylamine. This group of microorganisms also includes those capable of assimilating reduced one-carbon compounds by way of carbon dioxide using the ribulose bisphosphate pathway. These organisms should not be confused with methanogens which on the contrary produce methane as a by-product from various one-carbon compounds such as carbon dioxide. Some methylotrophs can degrade the greenhouse gas methane, and in this case they are called methanotrophs. The abundance, purity, and low price of methanol compared to commonly used sugars make methylotrophs competent organisms for production of amino acids, vitamins, recombinant proteins, single-cell proteins, co-enzymes and cytochromes.
Organohalide respiration (OHR) (previously named halorespiration or dehalorespiration) is the use of halogenated compounds as terminal electron acceptors in anaerobic respiration. Organohalide respiration can play a part in microbial biodegradation. The most common substrates are chlorinated aliphatics (PCE, TCE, chloroform) and chlorinated phenols. Organohalide-respiring bacteria are highly diverse. This trait is found in some Campylobacterota, Thermodesulfobacteriota, Chloroflexota (green nonsulfur bacteria), low G+C gram positive Clostridia, and ultramicrobacteria.

Methane monooxygenase (MMO) is an enzyme capable of oxidizing the C-H bond in methane as well as other alkanes. Methane monooxygenase belongs to the class of oxidoreductase enzymes.
Microbial metabolism is the means by which a microbe obtains the energy and nutrients it needs to live and reproduce. Microbes use many different types of metabolic strategies and species can often be differentiated from each other based on metabolic characteristics. The specific metabolic properties of a microbe are the major factors in determining that microbe's ecological niche, and often allow for that microbe to be useful in industrial processes or responsible for biogeochemical cycles.
In biology, syntrophy, syntrophism, or cross-feeding is the cooperative interaction between at least two microbial species to degrade a single substrate. This type of biological interaction typically involves the transfer of one or more metabolic intermediates between two or more metabolically diverse microbial species living in close proximity to each other. Thus, syntrophy can be considered an obligatory interdependency and a mutualistic metabolism between different microbial species, wherein the growth of one partner depends on the nutrients, growth factors, or substrates provided by the other(s).
Desulfatibacillum alkenivorans AK-01 is a specific strain of Desulfatibacillum alkenivorans.
Pseudomonas stutzeri is a Gram-negative soil bacterium that is motile, has a single polar flagellum, and is classified as bacillus, or rod-shaped. While this bacterium was first isolated from human spinal fluid, it has since been found in many different environments due to its various characteristics and metabolic capabilities. P. stutzeri is an opportunistic pathogen in clinical settings, although infections are rare. Based on 16S rRNA analysis, this bacterium has been placed in the P. stutzeri group, to which it lends its name.
Thauera butanivorans is a species of Gram-negative bacteria capable of growth on alkanes C2-C9, but not methane, as well as other organic substrates such as lactate, acetate, succinate and citrate. To grow on alkanes, this bacterium expresses a soluble di-iron monooxygenase closely related to soluble methane monooxygenase from methanotrophs.
Methylobacillus flagellatus is a species of aerobic bacteria.
Microbial biodegradation is the use of bioremediation and biotransformation methods to harness the naturally occurring ability of microbial xenobiotic metabolism to degrade, transform or accumulate environmental pollutants, including hydrocarbons, polychlorinated biphenyls (PCBs), polyaromatic hydrocarbons (PAHs), heterocyclic compounds, pharmaceutical substances, radionuclides and metals.

4-hydroxyphenylacetate 3-monooxygenase (EC 1.14.14.9) is an enzyme that catalyzes the chemical reaction

Lily Young is a distinguished professor of environmental microbiology at Rutgers New Brunswick. She is also a member of the administrative council at Rutgers University. She is the provost of Rutgers New Brunswick. She is a member of the Biotechnology Center for Agriculture and the Environment and has her academic appointment in the Department of Environmental Sciences.
Adsorbable organic halides (AOX) is a measure of the organic halogen load at a sampling site such as soil from a land fill, water, or sewage waste. The procedure measures chlorine, bromine, and iodine as equivalent halogens, but does not measure fluorine levels in the sample.
Bioremediation of petroleum contaminated environments is a process in which the biological pathways within microorganisms or plants are used to degrade or sequester toxic hydrocarbons, heavy metals, and other volatile organic compounds found within fossil fuels. Oil spills happen frequently at varying degrees along with all aspects of the petroleum supply chain, presenting a complex array of issues for both environmental and public health. While traditional cleanup methods such as chemical or manual containment and removal often result in rapid results, bioremediation is less labor-intensive, expensive, and averts chemical or mechanical damage. The efficiency and effectiveness of bioremediation efforts are based on maintaining ideal conditions, such as pH, RED-OX potential, temperature, moisture, oxygen abundance, nutrient availability, soil composition, and pollutant structure, for the desired organism or biological pathway to facilitate reactions. Three main types of bioremediation used for petroleum spills include microbial remediation, phytoremediation, and mycoremediation. Bioremediation has been implemented in various notable oil spills including the 1989 Exxon Valdez incident where the application of fertilizer on affected shoreline increased rates of biodegradation.

Bioremediation is the process of decontaminating polluted sites through the usage of either endogenous or external microorganism. In situ is a term utilized within a variety of fields meaning "on site" and refers to the location of an event. Within the context of bioremediation, in situ indicates that the location of the bioremediation has occurred at the site of contamination without the translocation of the polluted materials. Bioremediation is used to neutralize pollutants including Hydrocarbons, chlorinated compounds, nitrates, toxic metals and other pollutants through a variety of chemical mechanisms. Microorganism used in the process of bioremediation can either be implanted or cultivated within the site through the application of fertilizers and other nutrients. Common polluted sites targeted by bioremediation are groundwater/aquifers and polluted soils. Aquatic ecosystems affected by oil spills have also shown improvement through the application of bioremediation. The most notable cases being the Deepwater Horizon oil spill in 2010 and the Exxon Valdez oil spill in 1989. Two variations of bioremediation exist defined by the location where the process occurs. Ex situ bioremediation occurs at a location separate from the contaminated site and involves the translocation of the contaminated material. In situ occurs within the site of contamination In situ bioremediation can further be categorized by the metabolism occurring, aerobic and anaerobic, and by the level of human involvement.
Hydrocarbonoclastic bacteria are a heterogeneous group of prokaryotes which can degrade and utilize hydrocarbon compounds as source of carbon and energy. Despite being present in most of environments around the world, several of these specialized bacteria live in the sea and have been isolated from polluted seawater.
References
- 1 2 Joshua, C. J.; Dahl, R.; Benke, P. I.; Keasling, J. D. (2011). "Absence of Diauxie during Simultaneous Utilization of Glucose and Xylose by Sulfolobus acidocaldarius". J Bacteriol. 193 (6): 1293–1301. doi:10.1128/JB.01219-10. PMC 3067627 . PMID 21239580.
- ↑ Gulvik, C. A.; Buchan, A. (2013). "Simultaneous catabolism of plant-derived aromatic compounds results in enhanced growth for members of the Roseobacter lineage". Appl Environ Microbiol. 79 (12): 3716–3723. Bibcode:2013ApEnM..79.3716G. doi:10.1128/AEM.00405-13. PMC 3675927 . PMID 23563956.
- 1 2 Qin, Ke; Struckhoff, Garrett C.; Agrawal, Abinash; Shelley, Michael L.; Dong, Hailiang (2015-01-01). "Natural attenuation potential of tricholoroethene in wetland plant roots: Role of native ammonium-oxidizing microorganisms". Chemosphere. 119 (Supplement C): 971–977. Bibcode:2015Chmsp.119..971Q. doi:10.1016/j.chemosphere.2014.09.040. PMID 25303656.
- 1 2 3 4 5 Nzila, Alexis (2013-07-01). "Update on the cometabolism of organic pollutants by bacteria". Environmental Pollution. 178 (Supplement C): 474–482. Bibcode:2013EPoll.178..474N. doi:10.1016/j.envpol.2013.03.042. PMID 23570949.
- ↑ Beam, H. W.; Perry, J. J. (1973-03-01). "Co-metabolism as a factor in microbial degradation of cycloparaffinic hydrocarbons". Archiv für Mikrobiologie. 91 (1): 87–90. doi:10.1007/BF00409542. ISSN 0003-9276. PMID 4711459. S2CID 22727106.
- 1 2 3 4 Ryoo, D.; Shim, H.; Canada, K.; Barbieri, P.; Wood, T. K. (July 2000). "Aerobic degradation of tetrachloroethylene by toluene-o-xylene monooxygenase of Pseudomonas stutzeri OX1". Nature Biotechnology. 18 (7): 775–778. doi:10.1038/77344. ISSN 1087-0156. PMID 10888848. S2CID 19633783.
- 1 2 Li, Shanshan; Wang, Shan; Yan, Wei (2016). "Biodegradation of Methyl tert-Butyl Ether by Co-Metabolism with a Pseudomonas sp. Strain". International Journal of Environmental Research and Public Health. 13 (9): 883. doi: 10.3390/ijerph13090883 . ISSN 1661-7827. PMC 5036716 . PMID 27608032.
- ↑ Fraraccio, Serena; Strejcek, Michal; Dolinova, Iva; Macek, Tomas; Uhlik, Ondrej (2017-08-16). "Secondary compound hypothesis revisited: Selected plant secondary metabolites promote bacterial degradation of cis-1,2-dichloroethylene (cDCE)". Scientific Reports. 7 (1): 8406. Bibcode:2017NatSR...7.8406F. doi:10.1038/s41598-017-07760-1. ISSN 2045-2322. PMC 5559444 . PMID 28814712.
This article needs additional citations for verification .(June 2007) |