
Mendelian inheritance is a type of biological inheritance following the principles originally proposed by Gregor Mendel in 1865 and 1866, re-discovered in 1900 by Hugo de Vries and Carl Correns, and later popularized by William Bateson. These principles were initially controversial. When Mendel's theories were integrated with the Boveri–Sutton chromosome theory of inheritance by Thomas Hunt Morgan in 1915, they became the core of classical genetics. Ronald Fisher combined these ideas with the theory of natural selection in his 1930 book The Genetical Theory of Natural Selection, putting evolution onto a mathematical footing and forming the basis for population genetics within the modern evolutionary synthesis.

Chromosomal crossover, or crossing over, is the exchange of genetic material during sexual reproduction between two homologous chromosomes' non-sister chromatids that results in recombinant chromosomes. It is one of the final phases of genetic recombination, which occurs in the pachytene stage of prophase I of meiosis during a process called synapsis. Synapsis begins before the synaptonemal complex develops and is not completed until near the end of prophase I. Crossover usually occurs when matching regions on matching chromosomes break and then reconnect to the other chromosome.

Genetic recombination is the exchange of genetic material between different organisms which leads to production of offspring with combinations of traits that differ from those found in either parent. In eukaryotes, genetic recombination during meiosis can lead to a novel set of genetic information that can be further passed on from parents to offspring. Most recombination occurs naturally and can be classified into two types: (1) interchromosomal recombination, occurring through independent assortment of alleles whose loci are on different but homologous chromosomes ; & (2) intrachromosomal recombination, occurring through crossing over.
Genetic linkage is the tendency of DNA sequences that are close together on a chromosome to be inherited together during the meiosis phase of sexual reproduction. Two genetic markers that are physically near to each other are unlikely to be separated onto different chromatids during chromosomal crossover, and are therefore said to be more linked than markers that are far apart. In other words, the nearer two genes are on a chromosome, the lower the chance of recombination between them, and the more likely they are to be inherited together. Markers on different chromosomes are perfectly unlinked, although the penetrance of potentially deleterious alleles may be influenced by the presence of other alleles, and these other alleles may be located on other chromosomes than that on which a particular potentially deleterious allele is located.

A couple of homologous chromosomes, or homologs, are a set of one maternal and one paternal chromosome that pair up with each other inside a cell during fertilization. Homologs have the same genes in the same loci where they provide points along each chromosome which enable a pair of chromosomes to align correctly with each other before separating during meiosis. This is the basis for Mendelian inheritance which characterizes inheritance patterns of genetic material from an organism to its offspring parent developmental cell at the given time and area.
In population genetics, linkage disequilibrium (LD) is the non-random association of alleles at different loci in a given population. Loci are said to be in linkage disequilibrium when the frequency of association of their different alleles is higher or lower than expected if the loci were independent and associated randomly.
Forward genetics is a molecular genetics approach of determining the genetic basis responsible for a phenotype. Forward genetics provides an unbiased approach because it relies heavily on identifying the genes or genetic factors that cause a particular phenotype or trait of interest.

Non-Mendelian inheritance is any pattern in which traits do not segregate in accordance with Mendel's laws. These laws describe the inheritance of traits linked to single genes on chromosomes in the nucleus. In Mendelian inheritance, each parent contributes one of two possible alleles for a trait. If the genotypes of both parents in a genetic cross are known, Mendel's laws can be used to determine the distribution of phenotypes expected for the population of offspring. There are several situations in which the proportions of phenotypes observed in the progeny do not match the predicted values.
Genetics, a discipline of biology, is the science of heredity and variation in living organisms.
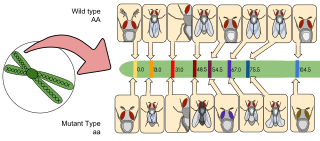
Gene mapping or genome mapping describes the methods used to identify the location of a gene on a chromosome and the distances between genes. Gene mapping can also describe the distances between different sites within a gene.
Genetic association is when one or more genotypes within a population co-occur with a phenotypic trait more often than would be expected by chance occurrence.
A tag SNP is a representative single nucleotide polymorphism (SNP) in a region of the genome with high linkage disequilibrium that represents a group of SNPs called a haplotype. It is possible to identify genetic variation and association to phenotypes without genotyping every SNP in a chromosomal region. This reduces the expense and time of mapping genome areas associated with disease, since it eliminates the need to study every individual SNP. Tag SNPs are useful in whole-genome SNP association studies in which hundreds of thousands of SNPs across the entire genome are genotyped.
Marker assisted selection or marker aided selection (MAS) is an indirect selection process where a trait of interest is selected based on a marker linked to a trait of interest, rather than on the trait itself. This process has been extensively researched and proposed for plant- and animal- breeding.
In genetics, a centimorgan or map unit (m.u.) is a unit for measuring genetic linkage. It is defined as the distance between chromosome positions for which the expected average number of intervening chromosomal crossovers in a single generation is 0.01. It is often used to infer distance along a chromosome. However, it is not a true physical distance.
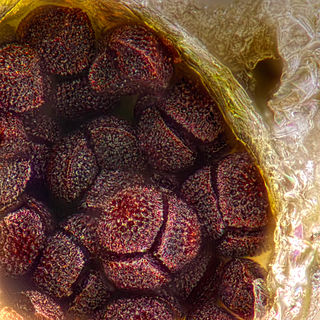
The tetrad is the four spores produced after meiosis of a yeast or other Ascomycota, Chlamydomonas or other alga, or a plant. After parent haploids mate, they produce diploids. Under appropriate environmental conditions, diploids sporulate and undergo meiosis. The meiotic products, spores, remain packaged in the parental cell body to produce the tetrad.
A doubled haploid (DH) is a genotype formed when haploid cells undergo chromosome doubling. Artificial production of doubled haploids is important in plant breeding.
A recombinant inbred strain or recombinant inbred line (RIL) is an organism with chromosomes that incorporate an essentially permanent set of recombination events between chromosomes inherited from two or more inbred strains. F1 and F2 generations are produced by intercrossing the inbred strains; pairs of the F2 progeny are then mated to establish inbred strains through long-term inbreeding.
Quantitative trait loci mapping or QTL mapping is the process of identifying genomic regions that potentially contain genes responsible for important economic, health or environmental characters. Mapping QTLs is an important activity that plant breeders and geneticists routinely use to associate potential causal genes with phenotypes of interest. Family-based QTL mapping is a variant of QTL mapping where multiple-families are used.
In genetics, the crossover value is the linked frequency of chromosomal crossover between two gene loci (markers). For a fixed set of genetic and environmental conditions, recombination in a particular region of a linkage structure (chromosome) tends to be constant and the same is then true for the crossover value which is used in the production of genetic maps.
This glossary of genetics and evolutionary biology is a list of definitions of terms and concepts used in the study of genetics and evolutionary biology, as well as sub-disciplines and related fields, with an emphasis on classical genetics, quantitative genetics, population biology, phylogenetics, speciation, and systematics. Overlapping and related terms can be found in Glossary of cellular and molecular biology, Glossary of ecology, and Glossary of biology.










