
In molecular biology, a riboswitch is a regulatory segment of a messenger RNA molecule that binds a small molecule, resulting in a change in production of the proteins encoded by the mRNA. Thus, an mRNA that contains a riboswitch is directly involved in regulating its own activity, in response to the concentrations of its effector molecule. The discovery that modern organisms use RNA to bind small molecules, and discriminate against closely related analogs, expanded the known natural capabilities of RNA beyond its ability to code for proteins, catalyze reactions, or to bind other RNA or protein macromolecules.
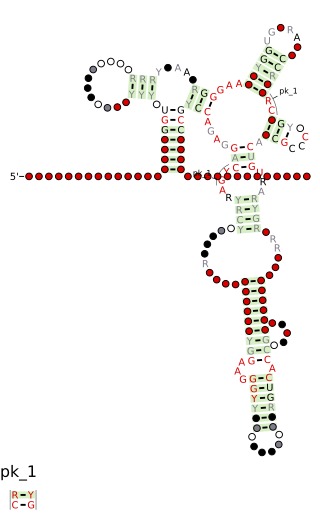
Cobalamin riboswitch is a cis-regulatory element which is widely distributed in 5' untranslated regions of vitamin B12 (Cobalamin) related genes in bacteria.

The YdaO/YuaA leader is a conserved RNA structure found upstream of the ydaO and yuaA genes in Bacillus subtilis and related genes in other bacteria. Its secondary structure and gene associations were predicted by bioinformatics.

The Lysine riboswitch is a metabolite binding RNA element found within certain messenger RNAs that serve as a precision sensor for the amino acid lysine. Allosteric rearrangement of mRNA structure is mediated by ligand binding, and this results in modulation of gene expression. Lysine riboswitch are most abundant in Bacillota and Gammaproteobacteria where they are found upstream of a number of genes involved in lysine biosynthesis, transport and catabolism. The lysine riboswitch has also been identified independently and called the L box.
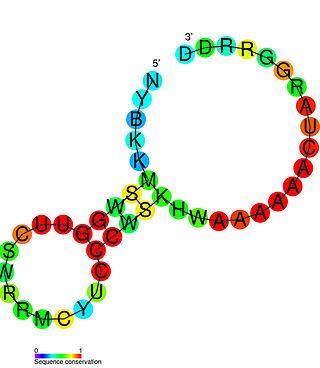
The PreQ1-I riboswitch is a cis-acting element identified in bacteria which regulates expression of genes involved in biosynthesis of the nucleoside queuosine (Q) from GTP. PreQ1 (pre-queuosine1) is an intermediate in the queuosine pathway, and preQ1 riboswitch, as a type of riboswitch, is an RNA element that binds preQ1. The preQ1 riboswitch is distinguished by its unusually small aptamer, compared to other riboswitches. Its atomic-resolution three-dimensional structure has been determined, with the PDB ID 2L1V.

The SAM-II riboswitch is an RNA element found predominantly in Alphaproteobacteria that binds S-adenosyl methionine (SAM). Its structure and sequence appear to be unrelated to the SAM riboswitch found in Gram-positive bacteria. This SAM riboswitch is located upstream of the metA and metC genes in Agrobacterium tumefaciens, and other methionine and SAM biosynthesis genes in other alpha-proteobacteria. Like the other SAM riboswitch, it probably functions to turn off expression of these genes in response to elevated SAM levels. A significant variant of SAM-II riboswitches was found in Pelagibacter ubique and related marine bacteria and called SAM-V. Also, like many structured RNAs, SAM-II riboswitches can tolerate long loops between their stems.

The SAM riboswitch is found upstream of a number of genes which code for proteins involved in methionine or cysteine biosynthesis in Gram-positive bacteria. Two SAM riboswitches in Bacillus subtilis that were experimentally studied act at the level of transcription termination control. The predicted secondary structure consists of a complex stem-loop region followed by a single stem-loop terminator region. An alternative and mutually exclusive form involves bases in the 3' segment of helix 1 with those in the 5' region of helix 5 to form a structure termed the anti-terminator form. When SAM is unbound, the anti-terminator sequence sequesters the terminator sequence so the terminator is unable to form, allowing the polymerase to read-through the downstream gene. When S-Adenosyl methionine (SAM) is bound to the aptamer, the anti-terminator is sequestered by an anti-anti-terminator; the terminator forms and terminates the transcription. However, many SAM riboswitches are likely to regulate gene expression at the level of translation.
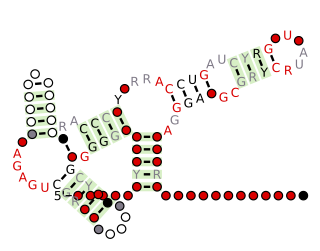
The TPP riboswitch, also known as the THI element and Thi-box riboswitch, is a highly conserved RNA secondary structure. It serves as a riboswitch that binds thiamine pyrophosphate (TPP) directly and modulates gene expression through a variety of mechanisms in archaea, bacteria and eukaryotes. TPP is the active form of thiamine (vitamin B1), an essential coenzyme synthesised by coupling of pyrimidine and thiazole moieties in bacteria. The THI element is an extension of a previously detected thiamin-regulatory element, the thi box, there is considerable variability in the predicted length and structures of the additional and facultative stem-loops represented in dark blue in the secondary structure diagram Analysis of operon structures has identified a large number of new candidate thiamin-regulated genes, mostly transporters, in various prokaryotic organisms. The x-ray crystal structure of the TPP riboswitch aptamer has been solved.

The ykkC/yxkD leader is a conserved RNA structure found upstream of the ykkC and yxkD genes in Bacillus subtilis and related genes in other bacteria. The function of this family is unclear for many years although it has been suggested that it may function to switch on efflux pumps and detoxification systems in response to harmful environmental molecules. The Thermoanaerobacter tengcongensis sequence AE013027 overlaps with that of purine riboswitch suggesting that the two riboswitches may work in conjunction to regulate the upstream gene which codes for TTE0584 (Q8RC62), a member of the permease family.

The Ykok leader or M-box is a Mg2+-sensing RNA structure that controls the expression of Magnesium ion transport proteins in bacteria. It is a distinct structure to the Magnesium responsive RNA element.

This family is a putative regulatory RNA structure that is found upstream of the ylbH gene in B. subtilis and related low GC Gram-positive bacteria.
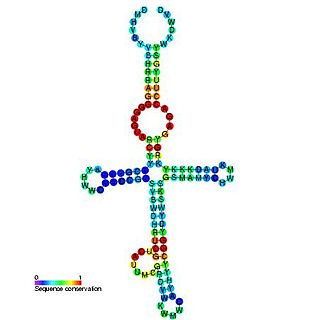
The yybP-ykoY leader RNA element was originally discovered in E. coli during a large scale screen and was named SraF. This family was later found to exist upstream of related families of protein genes in many bacteria, including the yybP and ykoY genes in B. subtilis. The specific functions of these proteins are unknown, but this structured RNA element may be involved in their genetic regulation as a riboswitch. The yybP-ykoY element was later proposed to be manganese-responsive after another associated family of genes, YebN/MntP, was shown to encode Mn2+ efflux pumps in several bacteria. Genetic data and a crystal structure confirmed that yybp-ykoY is a manganese riboswitch that directly binds Mn2+

PreQ1-II riboswitches form a class of riboswitches that specifically bind pre-queuosine1 (PreQ1), a precursor of the modified nucleoside queuosine. They are found in certain species of Streptococcus and Lactococcus, and were originally identified as a conserved RNA secondary structure called the "COG4708 motif". All known members of this riboswitch class appear to control members of COG4708 genes. These genes are predicted to encode membrane-bound proteins and have been proposed to be a transporter of preQ1, or a related metabolite, based on their association with preQ1-binding riboswitches. PreQ1-II riboswitches have no apparent similarities in sequence or structure to preQ1-I riboswitches, a previously discovered class of preQ1-binding riboswitches. PreQ1 thus joins S-adenosylmethionine as the second metabolite to be found that is the ligand of more than one riboswitch class.

Cyclic di-GMP is a second messenger used in signal transduction in a wide variety of bacteria. Cyclic di-GMP is not known to be used by archaea, and has only been observed in eukaryotes in Dictyostelium. The biological role of cyclic di-GMP was first uncovered when it was identified as an allosteric activator of a cellulose synthase found in Gluconacetobacter xylinus in order to produce microbial cellulose.

The fluoride riboswitch is a conserved RNA structure identified by bioinformatics in a wide variety of bacteria and archaea. These RNAs were later shown to function as riboswitches that sense fluoride ions. These "fluoride riboswitches" increase expression of downstream genes when fluoride levels are elevated, and the genes are proposed to help mitigate the toxic effects of very high levels of fluoride.

The glutamine riboswitch is a conserved RNA structure that was predicted by bioinformatics. It is present in a variety of lineages of cyanobacteria, as well as some phages that infect cyanobacteria. It is also found in DNA extracted from uncultivated bacteria living in the ocean that are presumably species of cyanobacteria.

The pfl RNA motif refers to a conserved RNA structure present in some bacteria and originally discovered using bioinformatics. pfl RNAs are consistently present in genomic locations that likely correspond to the 5' untranslated regions of protein-coding genes. This arrangement in bacteria is commonly associated with cis-regulatory elements. Moreover, they are in presumed 5' UTRs of multiple non-homologous genes, suggesting that they function only in these locations. Additional evidence of cis-regulatory function came from the observation that predicted rho-independent transcription terminators overlap pfl RNAs. This overlap suggests that the alternate secondary structures of pfl RNA and the transcription terminator stem-loops compete with each other, and this is a common mechanism for cis gene control in bacteria.

Tetrahydrofolate riboswitches are a class of homologous RNAs in certain bacteria that bind tetrahydrofolate (THF). It is almost exclusively located in the probable 5' untranslated regions of protein-coding genes, and most of these genes are known to encode either folate transporters or enzymes involved in folate metabolism. For these reasons it was inferred that the RNAs function as riboswitches. THF riboswitches are found in a variety of Bacillota, specifically the orders Clostridiales and Lactobacillales, and more rarely in other lineages of bacteria. The THF riboswitch was one of many conserved RNA structures found in a project based on comparative genomics. The 3-d structure of the tetrahydrofolate riboswitch has been solved by separate groups using X-ray crystallography. These structures were deposited into the Protein Data Bank under accessions 3SD1 and 3SUX, with other entries containing variants.

Cyclic di-GMP-II riboswitches form a class of riboswitches that specifically bind cyclic di-GMP, a second messenger used in multiple bacterial processes such as virulence, motility and biofilm formation. Cyclic di-GMP II riboswitches are structurally unrelated to cyclic di-GMP-I riboswitches, though they have the same function.
An RNA motif is a description of a group of RNAs that have a related structure. RNA motifs consist of a pattern of features within the primary sequence and secondary structure of related RNAs. Thus, it extends the concept of a sequence motif to include RNA secondary structure. The term "RNA motif" can refer both to the pattern and to the RNA sequences that match it.





















