 | |
| Names | |
|---|---|
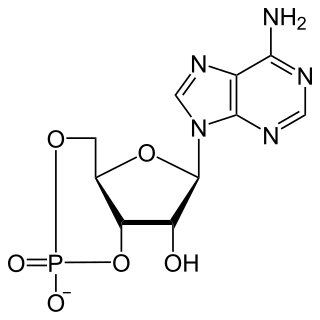
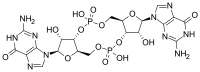
| Systematic IUPAC name (2R,3R,3aS,7aR,9R,10R,10aS,14aR)-2,9-Bis(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-3,5,10,12-tetrahydroxyoctahydro-2H,5H,7H,12H-5λ5,12λ5-difuro[3,2-d:3′,2′-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-5,12-dione | |
| Other names Cyclic diguanylate; 3',5'-Cyclic diguanylic acid; c-di-GMP; 5GP-5GP | |
| Identifiers | |
3D model (JSmol) | |
| ChemSpider | |
PubChem CID | |
CompTox Dashboard (EPA) | |
| |
| |
| Properties | |
| C20H24N10O14P2 | |
| Molar mass | 690.09 g/mol |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
Cyclic di-GMP (also called cyclic diguanylate and c-di-GMP ) is a second messenger used in signal transduction in a wide variety of bacteria. [1] Cyclic di-GMP is not known to be used by archaea, and has only been observed in eukaryotes in Dictyostelium . [2] The biological role of cyclic di-GMP was first uncovered when it was identified as an allosteric activator of a cellulose synthase found in Gluconacetobacter xylinus in order to produce microbial cellulose. [3]
In structure, it is a cycle containing only two guanine bases linked by ribose and phosphate.
Contact with surfaces increases c-di-GMP which increases transcription, translation, and post translation of exopolysaccharides (EPSs) and other extracellular polymeric substance matrix components (see the review by Jenal et al 2017). [4] In bacteria, certain signals are communicated by synthesizing or degrading cyclic di-GMP. Cyclic di-GMP is synthesized by proteins with diguanylate cyclase activity. These proteins typically have a characteristic GGDEF motif, which refers to a conserved sequence of five amino acids. Degradation of cyclic di-GMP is affected by proteins with phosphodiesterase activity. These proteins have either an EAL or an HD-GYP amino acid motif. Processes that are known to be regulated by cyclic di-GMP, at least in some organisms, include biofilm formation (such as EPS matrices found by Steiner et al 2013), [4] motility (especially the motile-to-sessile transition, see the review by Jenal et al 2017) [4] and virulence factor production.
Cyclic di-GMP levels are regulated using a variety of mechanisms. Many proteins with GGDEF, EAL or HD-GYP domains are found with other domains that can receive signals, such as PAS domains. Enzymes that degrade or synthesize cyclic di-GMP are believed to be localized to specific regions of the cell, where they influence receivers in a restricted space. [1] In Gluconacetobacter xylinus, c-di-GMP stimulates the polymerization of glucose into cellulose as a high affinity allosteric activator of the enzyme cellulose synthase. [3] Some diguanylate cyclase enzymes are allosterically inhibited by cyclic di-GMP.
Cyclic di-GMP levels regulate other processes via a number of mechanisms. The Gluconacetobacter xylinus cellulose synthase is allosterically stimulated by cyclic di-GMP, presenting a mechanism by which cyclic di-GMP can regulate cellulose synthase activity. The PilZ domain has been shown to bind cyclic di-GMP and is believed to be involved in cyclic di-GMP-dependent regulation, but the mechanism by which it does this is unknown. Recent structural studies of PilZ domains from two bacterial species have demonstrated that PilZ domains change conformation drastically upon binding to cyclic di-GMP. [5] [6] This leads to the strong inference that conformational changes in PilZ domains allow the activity of targeted effector proteins (such as cellulose synthase) to be regulated by cyclic di-GMP. Riboswitches called the cyclic di-GMP-I riboswitch and cyclic di-GMP-II riboswitch regulate gene expression in response to cyclic di-GMP concentrations in a variety of bacteria, but not all bacteria that are known to use cyclic di-GMP.
For a review of c-di-GMP roles in Caulobacter crescentus , Pseudomonas aeruginosa , Komagataeibacter xylinus /Gluconacetobacter xylinus, Myxococcus xanthus , Bdellovibrio bacteriovorus and Pseudomonas fluorescens see Jenal et al 2017. [4]