Biosynthesis
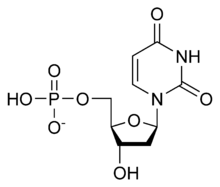
Deoxyuridine monophosphate (dUMP) is the deoxygenated form of uridine monophosphate (UMP), and is the precursor to deoxythymidine monophosphate (dTMP), a component of DNA nucleotide biosynthesis. [1] By replacing the hydroxyl group at the 2' carbon of ribose with a hydrogen, UMP becomes deoxygenated to dUMP.
The synthesis of deoxyuridine monophosphate (dUMP) is a multi-step process that begins with uridine monophosphate (UMP), the product of pyrimidine biosynthesis. [2] The enzyme nucleoside monophosphate kinase converts UMP and ATP to uridine diphosphate (UDP) and ADP.
In the presence of excess ATP, the enzyme ribonucleotide reductase initiates a chain reaction with UDP, which catalyzes the formation of deoxyuridine diphosphate (dUDP), which is then converted to deoxyuridine triphosphate (dUTP), then deoxyuridine monophosphate (dUMP) via the addition or removal of phosphate groups. [3]

Nucleotides are organic molecules composed of a nitrogenous base, a pentose sugar and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecules within all life-forms on Earth. Nucleotides are obtained in the diet and are also synthesized from common nutrients by the liver.
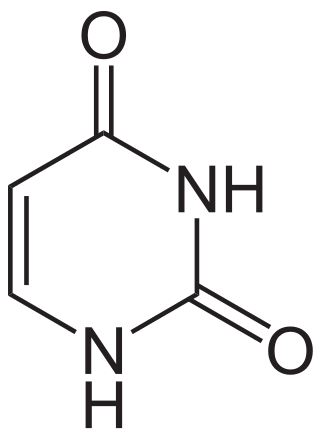
Uracil is one of the four nucleobases in the nucleic acid RNA. The others are adenine (A), cytosine (C), and guanine (G). In RNA, uracil binds to adenine via two hydrogen bonds. In DNA, the uracil nucleobase is replaced by thymine (T). Uracil is a demethylated form of thymine.
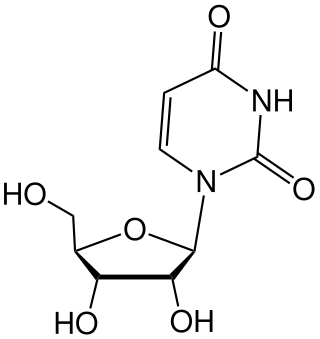
Uridine (symbol U or Urd) is a glycosylated pyrimidine analog containing uracil attached to a ribose ring (or more specifically, a ribofuranose) via a β-N1-glycosidic bond. The analog is one of the five standard nucleosides which make up nucleic acids, the others being adenosine, thymidine, cytidine and guanosine. The five nucleosides are commonly abbreviated to their symbols, U, A, dT, C, and G, respectively. However, thymidine is more commonly written as 'dT' ('d' represents 'deoxy') as it contains a 2'-deoxyribofuranose moiety rather than the ribofuranose ring found in uridine. This is because thymidine is found in deoxyribonucleic acid (DNA) and usually not in ribonucleic acid (RNA). Conversely, uridine is found in RNA and not DNA. The remaining three nucleosides may be found in both RNA and DNA. In RNA, they would be represented as A, C and G whereas in DNA they would be represented as dA, dC and dG.
The enzyme Uridine monophosphate synthase catalyses the formation of uridine monophosphate (UMP), an energy-carrying molecule in many important biosynthetic pathways. In humans, the gene that codes for this enzyme is located on the long arm of chromosome 3 (3q13).

In biochemistry, a ribonucleotide is a nucleotide containing ribose as its pentose component. It is considered a molecular precursor of nucleic acids. Nucleotides are the basic building blocks of DNA and RNA. Ribonucleotides themselves are basic monomeric building blocks for RNA. Deoxyribonucleotides, formed by reducing ribonucleotides with the enzyme ribonucleotide reductase (RNR), are essential building blocks for DNA. There are several differences between DNA deoxyribonucleotides and RNA ribonucleotides. Successive nucleotides are linked together via phosphodiester bonds.
A salvage pathway is a pathway in which a biological product is produced from intermediates in the degradative pathway of its own or a similar substance. The term often refers to nucleotide salvage in particular, in which nucleotides are synthesized from intermediates in their degradative pathway.
A nucleoside triphosphate is a nucleoside containing a nitrogenous base bound to a 5-carbon sugar, with three phosphate groups bound to the sugar. They are the molecular precursors of both DNA and RNA, which are chains of nucleotides made through the processes of DNA replication and transcription. Nucleoside triphosphates also serve as a source of energy for cellular reactions and are involved in signalling pathways.
In molecular biology, biosynthesis is a multi-step, enzyme-catalyzed process where substrates are converted into more complex products in living organisms. In biosynthesis, simple compounds are modified, converted into other compounds, or joined to form macromolecules. This process often consists of metabolic pathways. Some of these biosynthetic pathways are located within a single cellular organelle, while others involve enzymes that are located within multiple cellular organelles. Examples of these biosynthetic pathways include the production of lipid membrane components and nucleotides. Biosynthesis is usually synonymous with anabolism.
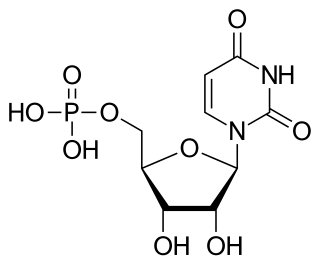
Uridine monophosphate (UMP), also known as 5′-uridylic acid, is a nucleotide that is used as a monomer in RNA. It is an ester of phosphoric acid with the nucleoside uridine. UMP consists of the phosphate group, the pentose sugar ribose, and the nucleobase uracil; hence, it is a ribonucleotide monophosphate. As a substituent or radical its name takes the form of the prefix uridylyl-. The deoxy form is abbreviated dUMP. Covalent attachment of UMP is called uridylylation.

Nucleic acid metabolism is a collective term that refers to the variety of chemical reactions by which nucleic acids are either synthesized or degraded. Nucleic acids are polymers made up of a variety of monomers called nucleotides. Nucleotide synthesis is an anabolic mechanism generally involving the chemical reaction of phosphate, pentose sugar, and a nitrogenous base. Degradation of nucleic acids is a catabolic reaction and the resulting parts of the nucleotides or nucleobases can be salvaged to recreate new nucleotides. Both synthesis and degradation reactions require multiple enzymes to facilitate the event. Defects or deficiencies in these enzymes can lead to a variety of diseases.

Orotidine 5'-phosphate decarboxylase or orotidylate decarboxylase is an enzyme involved in pyrimidine biosynthesis. It catalyzes the decarboxylation of orotidine monophosphate (OMP) to form uridine monophosphate (UMP). The function of this enzyme is essential to the de novo biosynthesis of the pyrimidine nucleotides uridine triphosphate, cytidine triphosphate, and thymidine triphosphate. OMP decarboxylase has been a frequent target for scientific investigation because of its demonstrated extreme catalytic efficiency and its usefulness as a selection marker for yeast strain engineering.
Pyrimidine biosynthesis occurs both in the body and through organic synthesis.

Orotate phosphoribosyltransferase (OPRTase) or orotic acid phosphoribosyltransferase is an enzyme involved in pyrimidine biosynthesis. It catalyzes the formation of orotidine 5'-monophosphate (OMP) from orotate and phosphoribosyl pyrophosphate. In yeast and bacteria, orotate phosphoribosyltransferase is an independent enzyme with a unique gene coding for the protein, whereas in mammals and other multicellular organisms, the catalytic function is carried out by a domain of the bifunctional enzyme UMP synthase (UMPS).

Orotic aciduria is a disease caused by an enzyme deficiency, resulting in a decreased ability to synthesize pyrimidines. It was the first described enzyme deficiency of the de novo pyrimidine synthesis pathway.
Purine metabolism refers to the metabolic pathways to synthesize and break down purines that are present in many organisms.

The enzyme UDP-glucose 4-epimerase, also known as UDP-galactose 4-epimerase or GALE, is a homodimeric epimerase found in bacterial, fungal, plant, and mammalian cells. This enzyme performs the final step in the Leloir pathway of galactose metabolism, catalyzing the reversible conversion of UDP-galactose to UDP-glucose. GALE tightly binds nicotinamide adenine dinucleotide (NAD+), a co-factor required for catalytic activity.
In enzymology, an UMP kinase is an enzyme that catalyzes the chemical reaction

Uridine-cytidine kinase 2 (UCK2) is an enzyme that in humans is encoded by the UCK2 gene.

UMP-CMP kinase is an enzyme that in humans is encoded by the CMPK1 gene.
David Sidney Feingold was an American biochemist.