
Adenylate cyclase is an enzyme with systematic name ATP diphosphate-lyase . It catalyzes the following reaction:
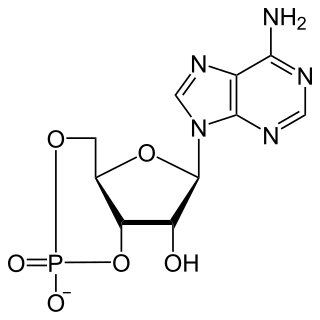
Cyclic adenosine monophosphate is a second messenger, or cellular signal occurring within cells, that is important in many biological processes. cAMP is a derivative of adenosine triphosphate (ATP) and used for intracellular signal transduction in many different organisms, conveying the cAMP-dependent pathway.

In biochemistry, a kinase is an enzyme that catalyzes the transfer of phosphate groups from high-energy, phosphate-donating molecules to specific substrates. This process is known as phosphorylation, where the high-energy ATP molecule donates a phosphate group to the substrate molecule. This transesterification produces a phosphorylated substrate and ADP. Conversely, it is referred to as dephosphorylation when the phosphorylated substrate donates a phosphate group and ADP gains a phosphate group. These two processes, phosphorylation and dephosphorylation, occur four times during glycolysis.

Adenosine monophosphate (AMP), also known as 5'-adenylic acid, is a nucleotide. AMP consists of a phosphate group, the sugar ribose, and the nucleobase adenine. It is an ester of phosphoric acid and the nucleoside adenosine. As a substituent it takes the form of the prefix adenylyl-.

In cell biology, protein kinase A (PKA) is a family of serine-threonine kinase whose activity is dependent on cellular levels of cyclic AMP (cAMP). PKA is also known as cAMP-dependent protein kinase. PKA has several functions in the cell, including regulation of glycogen, sugar, and lipid metabolism. It should not be confused with 5'-AMP-activated protein kinase.
Adenylate kinase is a phosphotransferase enzyme that catalyzes the interconversion of the various adenosine phosphates. By constantly monitoring phosphate nucleotide levels inside the cell, ADK plays an important role in cellular energy homeostasis.
CAMK, also written as CaMK or CCaMK, is an abbreviation for the Ca2+/calmodulin-dependent protein kinase class of enzymes. CAMKs are activated by increases in the concentration of intracellular calcium ions (Ca2+) and calmodulin. When activated, the enzymes transfer phosphates from ATP to defined serine or threonine residues in other proteins, so they are serine/threonine-specific protein kinases. Activated CAMK is involved in the phosphorylation of transcription factors and therefore, in the regulation of expression of responding genes. CAMK also works to regulate the cell life cycle (i.e. programmed cell death), rearrangement of the cell's cytoskeletal network, and mechanisms involved in the learning and memory of an organism.

Nucleoside-diphosphate kinases are enzymes that catalyze the exchange of terminal phosphate between different nucleoside diphosphates (NDP) and triphosphates (NTP) in a reversible manner to produce nucleotide triphosphates. Many NDP serve as acceptor while NTP are donors of phosphate group. The general reaction via ping-pong mechanism is as follows: XDP + YTP ←→ XTP + YDP. NDPK activities maintain an equilibrium between the concentrations of different nucleoside triphosphates such as, for example, when guanosine triphosphate (GTP) produced in the citric acid (Krebs) cycle is converted to adenosine triphosphate (ATP). Other activities include cell proliferation, differentiation and development, signal transduction, G protein-coupled receptor, endocytosis, and gene expression.

Guanosine monophosphate synthetase, also known as GMPS is an enzyme that converts xanthosine monophosphate to guanosine monophosphate.

Deoxycytidine kinase (dCK) is an enzyme which is encoded by the DCK gene in humans. dCK predominantly phosphorylates deoxycytidine (dC) and converts dC into deoxycytidine monophosphate. dCK catalyzes one of the initial steps in the nucleoside salvage pathway and has the potential to phosphorylate other preformed nucleosides, specifically deoxyadenosine (dA) and deoxyguanosine (dG), and convert them into their monophosphate forms. There has been recent biomedical research interest in investigating dCK's potential as a therapeutic target for different types of cancer.

Diphosphomevalonate decarboxylase (EC 4.1.1.33), most commonly referred to in scientific literature as mevalonate diphosphate decarboxylase, is an enzyme that catalyzes the chemical reaction

In enzymology, a guanylate kinase is an enzyme that catalyzes the chemical reaction
In enzymology, a nucleoside-triphosphate-adenylate kinase is an enzyme that catalyzes the chemical reaction
Phosphoribulokinase (PRK) (EC 2.7.1.19) is an essential photosynthetic enzyme that catalyzes the ATP-dependent phosphorylation of ribulose 5-phosphate (RuP) into ribulose 1,5-bisphosphate (RuBP), both intermediates in the Calvin Cycle. Its main function is to regenerate RuBP, which is the initial substrate and CO2-acceptor molecule of the Calvin Cycle. PRK belongs to the family of transferase enzymes, specifically those transferring phosphorus-containing groups (phosphotransferases) to an alcohol group acceptor. Along with ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBisCo), phosphoribulokinase is unique to the Calvin Cycle. Therefore, PRK activity often determines the metabolic rate in organisms for which carbon fixation is key to survival. Much initial work on PRK was done with spinach leaf extracts in the 1950s; subsequent studies of PRK in other photosynthetic prokaryotic and eukaryotic organisms have followed. The possibility that PRK might exist was first recognized by Weissbach et al. in 1954; for example, the group noted that carbon dioxide fixation in crude spinach extracts was enhanced by the addition of ATP. The first purification of PRK was conducted by Hurwitz and colleagues in 1956.
ATP + Mg2+ - D-ribulose 5-phosphate ADP + D-ribulose 1,5-bisphosphate
In enzymology, a polyphosphate kinase, or polyphosphate polymerase, is an enzyme that catalyzes the formation of polyphosphate from ATP, with chain lengths of up to a thousand or more orthophosphate moieties.

Pyruvate, phosphate dikinase, or PPDK is an enzyme in the family of transferases that catalyzes the chemical reaction

Uridine-cytidine kinase 2 (UCK2) is an enzyme that in humans is encoded by the UCK2 gene.

UMP-CMP kinase is an enzyme that in humans is encoded by the CMPK1 gene.
The Walker A and Walker B motifs are protein sequence motifs, known to have highly conserved three-dimensional structures. These were first reported in ATP-binding proteins by Walker and co-workers in 1982.

The prokaryotic riboflavin biosynthesis protein is a bifunctional enzyme found in bacteria that catalyzes the phosphorylation of riboflavin into flavin mononucleotide (FMN) and the adenylylation of FMN into flavin adenine dinucleotide (FAD). It consists of a C-terminal riboflavin kinase and an N-terminal FMN-adenylyltransferase. This bacterial protein is functionally similar to the monofunctional riboflavin kinases and FMN-adenylyltransferases of eukaryotic organisms, but only the riboflavin kinases are structurally homologous.

















