Nucleotides are organic molecules composed of a nitrogenous base, a pentose sugar and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecules within all life-forms on Earth. Nucleotides are obtained in the diet and are also synthesized from common nutrients by the liver.

Adenosine monophosphate (AMP), also known as 5'-adenylic acid, is a nucleotide. AMP consists of a phosphate group, the sugar ribose, and the nucleobase adenine. It is an ester of phosphoric acid and the nucleoside adenosine. As a substituent it takes the form of the prefix adenylyl-.

In biochemistry, a ribonucleotide is a nucleotide containing ribose as its pentose component. It is considered a molecular precursor of nucleic acids. Nucleotides are the basic building blocks of DNA and RNA. Ribonucleotides themselves are basic monomeric building blocks for RNA. Deoxyribonucleotides, formed by reducing ribonucleotides with the enzyme ribonucleotide reductase (RNR), are essential building blocks for DNA. There are several differences between DNA deoxyribonucleotides and RNA ribonucleotides. Successive nucleotides are linked together via phosphodiester bonds.
A nucleoside triphosphate is a nucleoside containing a nitrogenous base bound to a 5-carbon sugar, with three phosphate groups bound to the sugar. They are the molecular precursors of both DNA and RNA, which are chains of nucleotides made through the processes of DNA replication and transcription. Nucleoside triphosphates also serve as a source of energy for cellular reactions and are involved in signalling pathways.
Biosynthesis, i.e., chemical synthesis occurring in biological contexts, is a term most often referring to multi-step, enzyme-catalyzed processes where chemical substances absorbed as nutrients serve as enzyme substrates, with conversion by the living organism either into simpler or more complex products. Examples of biosynthetic pathways include those for the production of amino acids, lipid membrane components, and nucleotides, but also for the production of all classes of biological macromolecules, and of acetyl-coenzyme A, adenosine triphosphate, nicotinamide adenine dinucleotide and other key intermediate and transactional molecules needed for metabolism. Thus, in biosynthesis, any of an array of compounds, from simple to complex, are converted into other compounds, and so it includes both the catabolism and anabolism of complex molecules. Biosynthetic processes are often represented via charts of metabolic pathways. A particular biosynthetic pathway may be located within a single cellular organelle, while others involve enzymes that are located across an array of cellular organelles and structures.
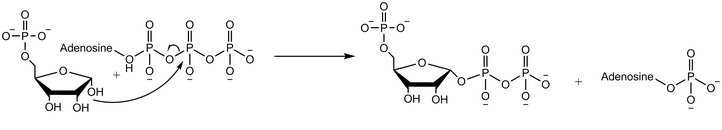
Adenine phosphoribosyltransferase (APRTase) is an enzyme encoded by the APRT gene, found in humans on chromosome 16. It is part of the Type I PRTase family and is involved in the nucleotide salvage pathway, which provides an alternative to nucleotide biosynthesis de novo in humans and most other animals. In parasitic protozoa such as giardia, APRTase provides the sole mechanism by which AMP can be produced. APRTase deficiency contributes to the formation of kidney stones (urolithiasis) and to potential kidney failure.
Purine nucleoside phosphorylase, PNP, PNPase or inosine phosphorylase is an enzyme that in humans is encoded by the NP gene. It catalyzes the chemical reaction
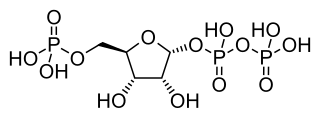
Phosphoribosyl pyrophosphate (PRPP) is a pentose phosphate. It is a biochemical intermediate in the formation of purine nucleotides via inosine-5-monophosphate, as well as in pyrimidine nucleotide formation. Hence it is a building block for DNA and RNA. The vitamins thiamine and cobalamin, and the amino acid tryptophan also contain fragments derived from PRPP. It is formed from ribose 5-phosphate (R5P) by the enzyme ribose-phosphate diphosphokinase:

Nucleic acid metabolism is a collective term that refers to the variety of chemical reactions by which nucleic acids are either synthesized or degraded. Nucleic acids are polymers made up of a variety of monomers called nucleotides. Nucleotide synthesis is an anabolic mechanism generally involving the chemical reaction of phosphate, pentose sugar, and a nitrogenous base. Degradation of nucleic acids is a catabolic reaction and the resulting parts of the nucleotides or nucleobases can be salvaged to recreate new nucleotides. Both synthesis and degradation reactions require multiple enzymes to facilitate the event. Defects or deficiencies in these enzymes can lead to a variety of diseases.

Ribose 5-phosphate (R5P) is both a product and an intermediate of the pentose phosphate pathway. The last step of the oxidative reactions in the pentose phosphate pathway is the production of ribulose 5-phosphate. Depending on the body's state, ribulose 5-phosphate can reversibly isomerize to ribose 5-phosphate. Ribulose 5-phosphate can alternatively undergo a series of isomerizations as well as transaldolations and transketolations that result in the production of other pentose phosphates as well as fructose 6-phosphate and glyceraldehyde 3-phosphate.
Phosphoribosylformylglycinamidine cyclo-ligase is the fifth enzyme in the de novo synthesis of purine nucleotides. It catalyzes the reaction to form 5-aminoimidazole ribotide (AIR) from formylglycinamidine-ribonucleotide FGAM. This reaction closes the ring and produces a 5-membered imidazole ring of the purine nucleus (AIR):

Orotate phosphoribosyltransferase (OPRTase) or orotic acid phosphoribosyltransferase is an enzyme involved in pyrimidine biosynthesis. It catalyzes the formation of orotidine 5'-monophosphate (OMP) from orotate and phosphoribosyl pyrophosphate. In yeast and bacteria, orotate phosphoribosyltransferase is an independent enzyme with a unique gene coding for the protein, whereas in mammals and other multicellular organisms, the catalytic function is carried out by a domain of the bifunctional enzyme UMP synthase (UMPS).
Purine metabolism refers to the metabolic pathways to synthesize and break down purines that are present in many organisms.

Phosphoribosylamine (PRA) is a biochemical intermediate in the formation of purine nucleotides via inosine-5-monophosphate, and hence is a building block for DNA and RNA. The vitamins thiamine and cobalamin also contain fragments derived from PRA.

Amidophosphoribosyltransferase (ATase), also known as glutamine phosphoribosylpyrophosphate amidotransferase (GPAT), is an enzyme responsible for catalyzing the conversion of 5-phosphoribosyl-1-pyrophosphate (PRPP) into 5-phosphoribosyl-1-amine (PRA), using the amine group from a glutamine side-chain. This is the committing step in de novo purine synthesis. In humans it is encoded by the PPAT gene. ATase is a member of the purine/pyrimidine phosphoribosyltransferase family.

Glycine—tRNA ligase also known as glycyl–tRNA synthetase is an enzyme that in humans is encoded by the GARS1 gene.

In molecular biology, the protein domain SAICAR synthase is an enzyme which catalyses a reaction to create SAICAR. In enzymology, this enzyme is also known as phosphoribosylaminoimidazolesuccinocarboxamide synthase. It is an enzyme that catalyzes the chemical reaction

5′-Phosphoribosyl-5-aminoimidazole is a biochemical intermediate in the formation of purine nucleotides via inosine-5-monophosphate, and hence is a building block for DNA and RNA. The vitamins thiamine and cobalamin also contain fragments derived from AIR. It is an intermediate in the adenine pathway and is synthesized from 5′-phosphoribosylformylglycinamidine by AIR synthetase.

Phosphoribosyl pyrophosphate synthetase-associated protein 2 is a protein that in humans is encoded by the PRPSAP2 gene.

Arts syndrome is a rare metabolic disorder that causes serious neurological problems in males due to a malfunction of the PRPP synthetase 1 enzyme. Arts Syndrome is part of a spectrum of PRPS-1 related disorders with reduced activity of the enzyme that includes Charcot–Marie–Tooth disease and X-linked non-syndromic sensorineural deafness.