Oxidative phosphorylation or electron transport-linked phosphorylation or terminal oxidation is the metabolic pathway in which cells use enzymes to oxidize nutrients, thereby releasing chemical energy in order to produce adenosine triphosphate (ATP). In eukaryotes, this takes place inside mitochondria. Almost all aerobic organisms carry out oxidative phosphorylation. This pathway is so pervasive because it releases more energy than alternative fermentation processes such as anaerobic glycolysis.
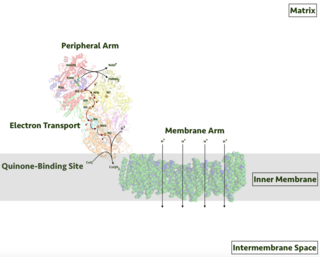
Respiratory complex I, EC 7.1.1.2 is the first large protein complex of the respiratory chains of many organisms from bacteria to humans. It catalyzes the transfer of electrons from NADH to coenzyme Q10 (CoQ10) and translocates protons across the inner mitochondrial membrane in eukaryotes or the plasma membrane of bacteria.

In biochemistry, flavin adenine dinucleotide (FAD) is a redox-active coenzyme associated with various proteins, which is involved with several enzymatic reactions in metabolism. A flavoprotein is a protein that contains a flavin group, which may be in the form of FAD or flavin mononucleotide (FMN). Many flavoproteins are known: components of the succinate dehydrogenase complex, α-ketoglutarate dehydrogenase, and a component of the pyruvate dehydrogenase complex.

Orotic acid is a pyrimidinedione and a carboxylic acid. Historically, it was believed to be part of the vitamin B complex and was called vitamin B13, but it is now known that it is not a vitamin.

The inner mitochondrial membrane (IMM) is the mitochondrial membrane which separates the mitochondrial matrix from the intermembrane space.
Pyrimidine biosynthesis occurs both in the body and through organic synthesis.

Electron-transferring-flavoprotein dehydrogenase is an enzyme that transfers electrons from electron-transferring flavoprotein in the mitochondrial matrix, to the ubiquinone pool in the inner mitochondrial membrane. It is part of the electron transport chain. The enzyme is found in both prokaryotes and eukaryotes and contains a flavin and FE-S cluster. In humans, it is encoded by the ETFDH gene. Deficiency in ETF dehydrogenase causes the human genetic disease multiple acyl-CoA dehydrogenase deficiency.
In enzymology, an orotate reductase (NADH) (EC 1.3.1.14) is an enzyme that catalyzes the chemical reaction
In enzymology, an orotate reductase (NADPH) (EC 1.3.1.15) is an enzyme that catalyzes the chemical reaction

NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13 is an enzyme that in humans is encoded by the NDUFA13 gene. The NDUFA13 protein is a subunit of NADH dehydrogenase (ubiquinone), which is located in the mitochondrial inner membrane and is the largest of the five complexes of the electron transport chain.

NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5 is an enzyme that in humans is encoded by the NDUFA5 gene. The NDUFA5 protein is a subunit of NADH dehydrogenase (ubiquinone), which is located in the mitochondrial inner membrane and is the largest of the five complexes of the electron transport chain.

NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9 is an enzyme that in humans is encoded by the NDUFB9 gene. NADH dehydrogenase (ubiquinone) 1 beta subcomplex subunit 9 is an accessory subunit of the NADH dehydrogenase (ubiquinone) complex, located in the mitochondrial inner membrane. It is also known as Complex I and is the largest of the five complexes of the electron transport chain.

NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8 is an enzyme that in humans is encoded by the NDUFA8 gene. The NDUFA8 protein is a subunit of NADH dehydrogenase (ubiquinone), which is located in the mitochondrial inner membrane and is the largest of the five complexes of the electron transport chain.

NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12 is an enzyme that in humans is encoded by the NDUFA12 gene. The NDUFA12 protein is a subunit of NADH dehydrogenase (ubiquinone), which is located in the mitochondrial inner membrane and is the largest of the five complexes of the electron transport chain. Mutations in subunits of NADH dehydrogenase (ubiquinone), also known as Complex I, frequently lead to complex neurodegenerative diseases such as Leigh's syndrome that result from mitochondrial complex I deficiency.

NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10 is an enzyme that in humans is encoded by the NDUFB10 gene. NADH dehydrogenase (ubiquinone) 1 beta subcomplex subunit 10 is an accessory subunit of the NADH dehydrogenase (ubiquinone) complex, located in the mitochondrial inner membrane. It is also known as Complex I and is the largest of the five complexes of the electron transport chain.

Miller syndrome, also known as Genée–Wiedemann syndrome, Wildervanck–Smith syndrome or postaxial acrofacial dysostosis, is an extremely rare genetic condition that manifests as craniofacial, limb and eye deformities. It is caused by a mutation in the DHODH gene. The incidence of the condition is not known, and nothing is known about its pathogenesis.

Class 2 dihydroorotate dehydrogenases is an enzyme with systematic name (S)-dihydroorotate:quinone oxidoreductase. This enzyme catalyses the electron transfer from dihydroorotate to a quinone :

NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa is a protein that in humans is encoded by the NDUFB3 gene. NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa is an accessory subunit of the NADH dehydrogenase (ubiquinone) complex, located in the mitochondrial inner membrane. It is also known as Complex I and is the largest of the five complexes of the electron transport chain. Mutations in this gene contribute to mitochondrial complex I deficiency.

NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 11 is an enzyme that in humans is encoded by the NDUFA11 gene. The NDUFA11 protein is a subunit of NADH dehydrogenase (ubiquinone), which is located in the mitochondrial inner membrane and is the largest of the five complexes of the electron transport chain Mutations in subunits of NADH dehydrogenase (ubiquinone), also known as Complex I, frequently lead to complex neurodegenerative diseases such as Leigh's syndrome. Mutations in this gene are associated with severe mitochondrial complex I deficiency.

S416 (GTPL-11164) is a drug which acts as a selective inhibitor of the enzyme dihydroorotate dehydrogenase (DHODH). This enzyme is involved in the synthesis of pyrimidine nucleosides in the body, which are required for the synthesis of DNA and RNA. This is an important rate-limiting step in the replication of viruses, and so DHODH inhibitors may have applications as broad-spectrum antiviral drugs. In tests in vitro, S416 was found to have antiviral activity against a range of pathogenic RNA viruses including influenza, Zika virus, Ebola virus and SARS-CoV-2.