
Nucleotides are organic molecules consisting of a nucleoside and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecules within all life-forms on Earth. Nucleotides are obtained in the diet and are also synthesized from common nutrients by the liver.

In molecular biology and genetics, translation is the process in which ribosomes in the cytoplasm or endoplasmic reticulum synthesize proteins after the process of transcription of DNA to RNA in the cell's nucleus. The entire process is called gene expression.
A salvage pathway is a pathway in which a biological product is produced from intermediates in the degradative pathway of its own or a similar substance. The term often refers to nucleotide salvage in particular, in which nucleotides are synthesized from intermediates in their degradative pathway.
A nucleoside triphosphate is a molecule containing a nitrogenous base bound to a 5-carbon sugar, with three phosphate groups bound to the sugar. It is an example of a nucleotide. They are the molecular precursors of both DNA and RNA, which are chains of nucleotides made through the processes of DNA replication and transcription. Nucleoside triphosphates also serve as a source of energy for cellular reactions and are involved in signalling pathways.
The Shine–Dalgarno (SD) sequence is a ribosomal binding site in bacterial and archaeal messenger RNA, generally located around 8 bases upstream of the start codon AUG. The RNA sequence helps recruit the ribosome to the messenger RNA (mRNA) to initiate protein synthesis by aligning the ribosome with the start codon. Once recruited, tRNA may add amino acids in sequence as dictated by the codons, moving downstream from the translational start site.

Nucleoside-diphosphate kinases are enzymes that catalyze the exchange of terminal phosphate between different nucleoside diphosphates (NDP) and triphosphates (NTP) in a reversible manner to produce nucleotide triphosphates. Many NDP serve as acceptor while NTP are donors of phosphate group. The general reaction via ping-pong mechanism is as follows: XDP + YTP ←→ XTP + YDP. NDPK activities maintain an equilibrium between the concentrations of different nucleoside triphosphates such as, for example, when guanosine triphosphate (GTP) produced in the citric acid (Krebs) cycle is converted to adenosine triphosphate (ATP). Other activities include cell proliferation, differentiation and development, signal transduction, G protein-coupled receptor, endocytosis, and gene expression.
Bacterial translation is the process by which messenger RNA is translated into proteins in bacteria.
The Kozak consensus sequence is a nucleic acid motif that functions as the protein translation initiation site in most eukaryotic mRNA transcripts. Regarded as the optimum sequence for initiating translation in eukaryotes, the sequence is an integral aspect of protein regulation and overall cellular health as well as having implications in human disease. It ensures that a protein is correctly translated from the genetic message, mediating ribosome assembly and translation initiation. A wrong start site can result in non-functional proteins. As it has become more studied, expansions of the nucleotide sequence, bases of importance, and notable exceptions have arisen. The sequence was named after the scientist who discovered it, Marilyn Kozak. Kozak discovered the sequence through a detailed analysis of DNA genomic sequences.

The trp operon is an operon-a group of genes that is used, or transcribed, together—that codes for the components for production of tryptophan. The trp operon is present in many bacteria, but was first characterized in Escherichia coli. The operon is regulated so that, when tryptophan is present in the environment, the genes for tryptophan synthesis are not expressed. It was an important experimental system for learning about gene regulation, and is commonly used to teach gene regulation.

Nucleic acid metabolism is the process by which nucleic acids are synthesized and degraded. Nucleic acids are the polymers of nucleotides. Nucleotide synthesis is an anabolic mechanism generally involving the chemical reaction of phosphate, pentose sugar, and a nitrogenous base. Destruction of nucleic acid is a catabolic reaction. Additionally, parts of the nucleotides or nucleobases can be salvaged to recreate new nucleotides. Both synthesis and degradation reactions require enzymes to facilitate the event. Defects or deficiencies in these enzymes can lead to a variety of diseases.
Pyrimidine biosynthesis occurs both in the body and through organic synthesis.
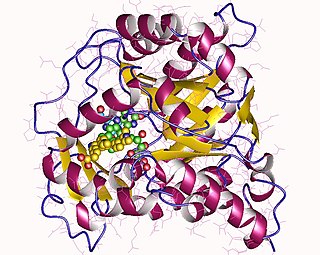
Dihydroorotate dehydrogenase (DHODH) is an enzyme that in humans is encoded by the DHODH gene on chromosome 16. The protein encoded by this gene catalyzes the fourth enzymatic step, the ubiquinone-mediated oxidation of dihydroorotate to orotate, in de novo pyrimidine biosynthesis. This protein is a mitochondrial protein located on the outer surface of the inner mitochondrial membrane (IMM). Inhibitors of this enzyme are used to treat autoimmune diseases such as rheumatoid arthritis.
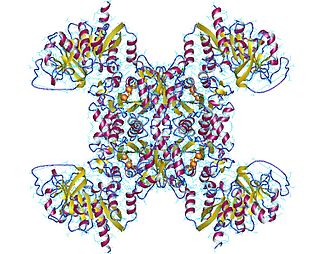
CTP synthase is an enzyme involved in pyrimidine biosynthesis that interconverts UTP and CTP.

Bacterial transcription is the process in which a segment of bacterial DNA is copied into a newly synthesized strand of messenger RNA (mRNA) with use of the enzyme RNA polymerase.
Translational regulation refers to the control of the levels of protein synthesized from its mRNA. This regulation is vastly important to the cellular response to stressors, growth cues, and differentiation. In comparison to transcriptional regulation, it results in much more immediate cellular adjustment through direct regulation of protein concentration. The corresponding mechanisms are primarily targeted on the control of ribosome recruitment on the initiation codon, but can also involve modulation of peptide elongation, termination of protein synthesis, or ribosome biogenesis. While these general concepts are widely conserved, some of the finer details in this sort of regulation have been proven to differ between prokaryotic and eukaryotic organisms.

FourU thermometers are a class of non-coding RNA thermometers found in Salmonella. They are named 'FourU' due to the four highly conserved uridine nucleotides found directly opposite the Shine-Dalgarno sequence on hairpin II (pictured). RNA thermometers such as FourU control regulation of temperature via heat shock proteins in many prokaryotes. FourU thermometers are relatively small RNA molecules, only 57 nucleotides in length, and have a simple two-hairpin structure.
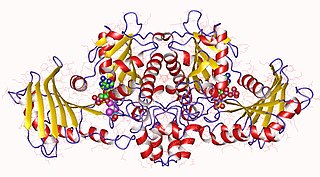
In molecular biology, acetate kinase (EC 2.7.2.1), which is predominantly found in micro-organisms, facilitates the production of acetyl-CoA by phosphorylating acetate in the presence of ATP and a divalent cation. Short-chain fatty acids (SCFAs) play a major role in carbon cycle and can be utilized as a source of carbon and energy by bacteria. Salmonella typhimurium propionate kinase (StTdcD) catalyzes reversible transfer of the γ-phosphate of ATP to propionate during l-threonine degradation to propionate. Kinetic analysis revealed that StTdcD possesses broad ligand specificity and could be activated by various SCFAs (propionate>acetate≈butyrate), nucleotides (ATP≈GTP>CTP≈TTP; dATP>dGTP>dCTP) and metal ions (Mg2+≈Mn2+>Co2+). Inhibition of StTdcD by tricarboxylic acid (TCA) cycle intermediates such as citrate, succinate, α-ketoglutarate and malate suggests that the enzyme could be under plausible feedback regulation. Crystal structures of StTdcD bound to PO4 (phosphate), AMP, ATP, Ap4 (adenosine tetraphosphate), GMP, GDP, GTP, CMP and CTP revealed that binding of nucleotide mainly involves hydrophobic interactions with the base moiety and could account for the broad biochemical specificity observed between the enzyme and nucleotides. Modelling and site-directed mutagenesis studies suggest Ala88 to be an important residue involved in determining the rate of catalysis with SCFA substrates. Molecular dynamics simulations on monomeric and dimeric forms of StTdcD revealed plausible open and closed states, and also suggested role for dimerization in stabilizing segment 235-290 involved in interfacial interactions and ligand binding. Observation of an ethylene glycol molecule bound sufficiently close to the γ-phosphate in StTdcD complexes with triphosphate nucleotides supports direct in-line phosphoryl transfer. The enzyme is important in the process of glycolysis, enzyme levels being increased in the presence of excess glucose. The growth of a bacterial mutant lacking acetate kinase has been shown to be inhibited by glucose, suggesting that the enzyme is involved in excretion of excess carbohydrate. A related enzyme, butyrate kinase, facilitates the formation of butyryl-CoA by phosphorylating butyrate in the presence of ATP to form butyryl phosphate.
The gua operon is responsible for regulating the synthesis of guanosine mono phosphate (GMP), a purine nucleotide, from inosine monophosphate. It consists of two structural genes guaB (encodes for IMP dehydrogenase or and guaA apart from the promoter and operator region.
In molecular biology, the PyrG leader is a cis-regulatory RNA element found at the 5' of the PyrG mRNA. The PyrG gene encodes a CTP synthase, which is involved in pyrimidine biosynthesis. The PyrG leader regulates expression of PyrG, PyrG can form into two different hairpin structures, a terminator or an anti-terminator. Under low CTP conditions, guanine (G) residues are incorporated at a specific site within the PyrG leader, these allow base-pairing with a uracil (U)-rich region and the formation of an anti-terminator loop, this results in increased expression of PyrG. Under high CTP conditions the guanines are not added, the anti-terminator loop cannot form and instead a terminator loop is formed, preventing further PyrG expression.
In molecular biology, the PyrC leader is a cis-regulatory RNA element found at the 5' of the PyrC mRNA in Enterobacteria. The PyrC gene encodes Dihydroorotase, an enzyme involved in pyrimidine biosynthesis. The PyrC leader regulates expression of PyrC. Translation initiation can occur at four different sites within this leader sequence, under high CTP conditions the translation initiation site is upstream of that used under low CTP conditions, additional cytosine residues are incorporated into the mRNA resulting in the formation of an RNA hairpin. This hairpin blocks ribosome-binding at the Shine-Dalgarno sequence, and therefore blocks expression of PyrC. Under low CTP conditions the initiation site is further downstream and does not result in hairpin formation, so the ribosome can bind to the Shine-Dalgarno sequence and PyrC is expressed.









