
Oxidative phosphorylation or electron transport-linked phosphorylation or terminal oxidation is the metabolic pathway in which cells use enzymes to oxidize nutrients, thereby releasing chemical energy in order to produce adenosine triphosphate (ATP). In eukaryotes, this takes place inside mitochondria. Almost all aerobic organisms carry out oxidative phosphorylation. This pathway is so pervasive because it releases more energy than alternative fermentation processes such as anaerobic glycolysis.
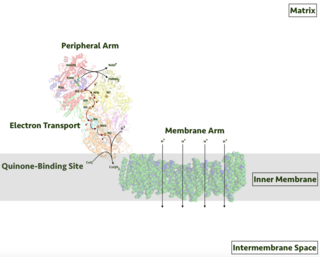
Respiratory complex I, EC 7.1.1.2 is the first large protein complex of the respiratory chains of many organisms from bacteria to humans. It catalyzes the transfer of electrons from NADH to coenzyme Q10 (CoQ10) and translocates protons across the inner mitochondrial membrane in eukaryotes or the plasma membrane of bacteria.
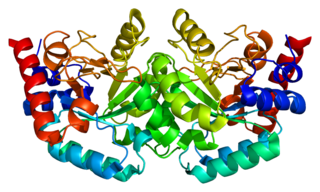
The enzyme Uridine monophosphate synthase catalyses the formation of uridine monophosphate (UMP), an energy-carrying molecule in many important biosynthetic pathways. In humans, the gene that codes for this enzyme is located on the long arm of chromosome 3 (3q13).
DNA gyrase, or simply gyrase, is an enzyme within the class of topoisomerase and is a subclass of Type II topoisomerases that reduces topological strain in an ATP dependent manner while double-stranded DNA is being unwound by elongating RNA-polymerase or by helicase in front of the progressing replication fork. It is the only known enzyme to actively contribute negative supercoiling to DNA, while it also is capable of relaxing positive supercoils. It does so by looping the template to form a crossing, then cutting one of the double helices and passing the other through it before releasing the break, changing the linking number by two in each enzymatic step. This process occurs in bacteria, whose single circular DNA is cut by DNA gyrase and the two ends are then twisted around each other to form supercoils. Gyrase is also found in eukaryotic plastids: it has been found in the apicoplast of the malarial parasite Plasmodium falciparum and in chloroplasts of several plants. Bacterial DNA gyrase is the target of many antibiotics, including nalidixic acid, novobiocin, albicidin, and ciprofloxacin.

Isocitrate dehydrogenase (IDH) (EC 1.1.1.42) and (EC 1.1.1.41) is an enzyme that catalyzes the oxidative decarboxylation of isocitrate, producing alpha-ketoglutarate (α-ketoglutarate) and CO2. This is a two-step process, which involves oxidation of isocitrate (a secondary alcohol) to oxalosuccinate (a ketone), followed by the decarboxylation of the carboxyl group beta to the ketone, forming alpha-ketoglutarate. In humans, IDH exists in three isoforms: IDH3 catalyzes the third step of the citric acid cycle while converting NAD+ to NADH in the mitochondria. The isoforms IDH1 and IDH2 catalyze the same reaction outside the context of the citric acid cycle and use NADP+ as a cofactor instead of NAD+. They localize to the cytosol as well as the mitochondrion and peroxisome.
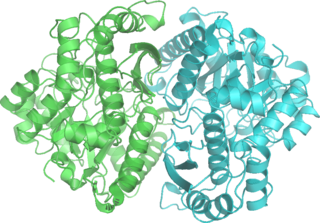
Phosphopyruvate hydratase, usually known as enolase, is a metalloenzyme (EC 4.2.1.11) that catalyses the conversion of 2-phosphoglycerate (2-PG) to phosphoenolpyruvate (PEP), the ninth and penultimate step of glycolysis. The chemical reaction is:

The TIM barrel, also known as an alpha/beta barrel, is a conserved protein fold consisting of eight alpha helices (α-helices) and eight parallel beta strands (β-strands) that alternate along the peptide backbone. The structure is named after triose-phosphate isomerase, a conserved metabolic enzyme. TIM barrels are ubiquitous, with approximately 10% of all enzymes adopting this fold. Further, five of seven enzyme commission (EC) enzyme classes include TIM barrel proteins. The TIM barrel fold is evolutionarily ancient, with many of its members possessing little similarity today, instead falling within the twilight zone of sequence similarity.
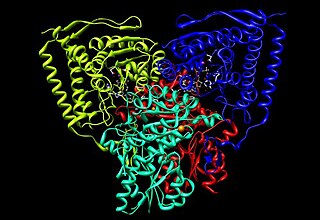
Pyruvate dehydrogenase is an enzyme that catalyzes the reaction of pyruvate and a lipoamide to give the acetylated dihydrolipoamide and carbon dioxide. The conversion requires the coenzyme thiamine pyrophosphate.
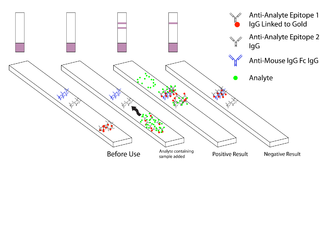
Malaria antigen detection tests are a group of commercially available rapid diagnostic tests of the rapid antigen test type that allow quick diagnosis of malaria by people who are not otherwise skilled in traditional laboratory techniques for diagnosing malaria or in situations where such equipment is not available. There are currently over 20 such tests commercially available. The first malaria antigen suitable as target for such a test was a soluble glycolytic enzyme Glutamate dehydrogenase. None of the rapid tests are currently as sensitive as a thick blood film, nor as cheap. A major drawback in the use of all current dipstick methods is that the result is essentially qualitative. In many endemic areas of tropical Africa, however, the quantitative assessment of parasitaemia is important, as a large percentage of the population will test positive in any qualitative assay.
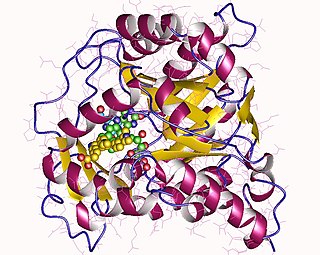
Dihydroorotate dehydrogenase (DHODH) is an enzyme that in humans is encoded by the DHODH gene on chromosome 16. The protein encoded by this gene catalyzes the fourth enzymatic step, the ubiquinone-mediated oxidation of dihydroorotate to orotate, in de novo pyrimidine biosynthesis. This protein is a mitochondrial protein located on the outer surface of the inner mitochondrial membrane (IMM). Inhibitors of this enzyme are used to treat autoimmune diseases such as rheumatoid arthritis.

6-Phosphogluconolactonase (EC 3.1.1.31, 6PGL, PGLS, systematic name 6-phospho-D-glucono-1,5-lactone lactonohydrolase) is a cytosolic enzyme found in all organisms that catalyzes the hydrolysis of 6-phosphogluconolactone to 6-phosphogluconic acid in the oxidative phase of the pentose phosphate pathway:
In enzymology, a quinoprotein glucose dehydrogenase is an enzyme that catalyzes the chemical reaction
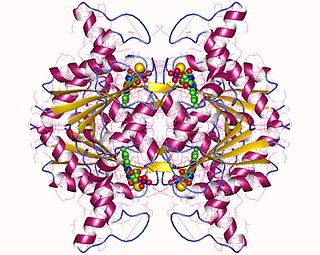
In enzymology, a NAD(P)H dehydrogenase (quinone) (EC 1.6.5.2) is an enzyme that catalyzes the chemical reaction

Ribose-5-phosphate isomerase (Rpi) encoded by the RPIA gene is an enzyme that catalyzes the conversion between ribose-5-phosphate (R5P) and ribulose-5-phosphate (Ru5P). It is a member of a larger class of isomerases which catalyze the interconversion of chemical isomers. It plays a vital role in biochemical metabolism in both the pentose phosphate pathway and the Calvin cycle. The systematic name of this enzyme class is D-ribose-5-phosphate aldose-ketose-isomerase.

Cystathionine beta-lyase, also commonly referred to as CBL or β-cystathionase, is an enzyme that primarily catalyzes the following α,β-elimination reaction
In enzymology, a 2-isopropylmalate synthase (EC 2.3.3.13) is an enzyme that catalyzes the chemical reaction

In enzymology, a malate synthase (EC 2.3.3.9) is an enzyme that catalyzes the chemical reaction

Fumarate reductase (quinol) (EC 1.3.5.4, QFR,FRD, menaquinol-fumarate oxidoreductase, quinol:fumarate reductase) is an enzyme with systematic name succinate:quinone oxidoreductase. This enzyme catalyzes the following chemical reaction:

NADH:ubiquinone reductase (non-electrogenic) (EC 1.6.5.9, NDH-2, ubiquinone reductase, coenzyme Q reductase, dihydronicotinamide adenine dinucleotide-coenzyme Q reductase, DPNH-coenzyme Q reductase, DPNH-ubiquinone reductase, NADH-coenzyme Q oxidoreductase, NADH-coenzyme Q reductase, NADH-CoQ oxidoreductase, NADH-CoQ reductase) is an enzyme with systematic name NADH:ubiquinone oxidoreductase. This enzyme catalyses the following chemical reaction:

NDH-2, also known as type II NADH:quinone oxidoreductase or alternative NADH dehydrogenase, is an enzyme which catalyzes the electron transfer from NADH to a quinone, being part of the electron transport chain. NDH-2 are peripheral membrane protein, functioning as dimers in vivo, with approximately 45 KDa per subunit and a single FAD as their cofactor.



















