Related Research Articles
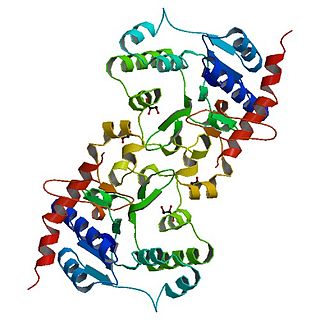
Glycogenin is an enzyme involved in converting glucose to glycogen. It acts as a primer, by polymerizing the first few glucose molecules, after which other enzymes take over. It is a homodimer of 37-kDa subunits and is classified as a glycosyltransferase.
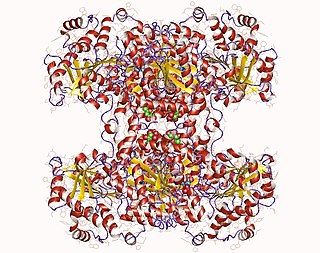
Glycogen synthase is a key enzyme in glycogenesis, the conversion of glucose into glycogen. It is a glycosyltransferase that catalyses the reaction of UDP-glucose and n to yield UDP and n+1.

Glycosyltransferases are enzymes that establish natural glycosidic linkages. They catalyze the transfer of saccharide moieties from an activated nucleotide sugar to a nucleophilic glycosyl acceptor molecule, the nucleophile of which can be oxygen- carbon-, nitrogen-, or sulfur-based.

Sialyltransferases are enzymes that transfer sialic acid to nascent oligosaccharide. Each sialyltransferase is specific for a particular sugar substrate. Sialyltransferases add sialic acid to the terminal portions of the sialylated glycolipids (gangliosides) or to the N- or O-linked sugar chains of glycoproteins.

Galactosyltransferase is a type of glycosyltransferase which catalyzes the transfer of galactose. An example is B-N-acetylglucosaminyl-glycopeptide b-1,4-galactosyltransferase.

Curdlan is a water-insoluble linear beta-1,3-glucan, a high-molecular-weight polymer of glucose. Curdlan consists of β-(1,3)-linked glucose residues and forms elastic gels upon heating in aqueous suspension. It was reported to be produced by Alcaligenes faecalis var. myxogenes. Subsequently, the taxonomy of this non-pathogenic curdlan-producing bacterium has been reclassified as Agrobacterium species.
N-acetyllactosamine synthase is a galactosyltransferase enzyme. It is a component of lactose synthase This enzyme modifies the connection between two molecule UDP-galactose and N-actyl-D-glucosamine and generates two different molecules UDP and N-acetyllactosamine as products. The main function of the enzyme is associated with the biosynthesis of glycoproteins and glycolipids in both human and animals. In human, the activity of this enzyme can be found in Golgi apparatus.
In enzymology, a 1,3-beta-D-glucan phosphorylase is an enzyme that catalyzes the chemical reaction
In enzymology, an alpha-1,3-glucan synthase is an enzyme that catalyzes the chemical reaction
In enzymology, a cellulose synthase (GDP-forming) is an enzyme that catalyzes the chemical reaction

The UDP-forming form of cellulose synthase is the main enzyme that produces cellulose. Systematically, it is known as UDP-glucose:(1→4)-β-D-glucan 4-β-D-glucosyltransferase in enzymology. It catalyzes the chemical reaction:
In enzymology, a ganglioside galactosyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a globotriaosylceramide 3-beta-N-acetylgalactosaminyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a glucosaminylgalactosylglucosylceramide beta-galactosyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a lactosylceramide 1,3-N-anning-beta-D-glrofelucosaminyltlolferase is an enzyme that catalyzes the chemical reaction
In enzymology, a lactosylceramide beta-1,3-galactosyltransferase is an enzyme that catalyzes the chemical reaction

In enzymology, a starch synthase is an enzyme that catalyzes the chemical reaction
In enzymology, a sucrose-1,6-alpha-glucan 3(6)-alpha-glucosyltransferase is an enzyme that catalyzes the chemical reaction
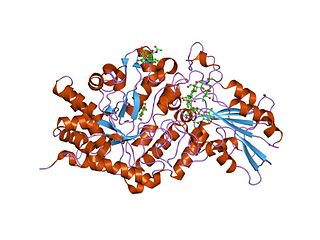
In molecular biology, glycoside hydrolase family 3 is a family of glycoside hydrolases. Glycoside hydrolases EC 3.2.1. are a widespread group of enzymes that hydrolyse the glycosidic bond between two or more carbohydrates, or between a carbohydrate and a non-carbohydrate moiety. A classification system for glycoside hydrolases, based on sequence similarity, has led to the definition of over 100 different families. This classification is available on the CAZy web site, and also discussed at CAZypedia, an online encyclopedia of carbohydrate active enzymes.

Glucanases are enzymes that break down large polysaccharides via hydrolysis. The product of the hydrolysis reaction is called a glucan, a linear polysaccharide made of up to 1200 glucose monomers, held together with glycosidic bonds. Glucans are abundant in the endosperm cell walls of cereals such as barley, rye, sorghum, rice, and wheat. Glucanases are also referred to as lichenases, hydrolases, glycosidases, glycosyl hydrolases, and/or laminarinases. Many types of glucanases share similar amino acid sequences but vastly different substrates. Of the known endo-glucanases, 1,3-1,4-β-glucanase is considered the most active.
References
- 1 2 3 KFLGG, ENZYME: 2.4.1.34
- ↑ Campbell JA, Davies GJ, Bulone V, Henrissat B (September 1997). "A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities". Biochem. J. 326 (3): 929–39. doi:10.1042/bj3260929u. PMC 1218753 . PMID 9334165.
- ↑ Karnezis T, Epa VC, Stone BA, Stanisich VA (2003). "Topological characterization of an inner membrane (1→3)-beta-D-glucan (curdlan) synthase from Agrobacterium sp. strain ATCC31749". Glycobiology. 13 (10): 693–706. doi: 10.1093/glycob/cwg093 . PMID 12851288.
- ↑ Mio T, Adachi-Shimizu M, Tachibana Y, Tabuchi H, Inoue SB, Yabe T, Yamada-Okabe T, Arisawa M, Watanabe T, Yamada-Okabe H (July 1997). "Cloning of the Candida albicans homolog of Saccharomyces cerevisiae GSC1/FKS1 and its involvement in beta-1,3-glucan synthesis". J. Bacteriol. 179 (13): 4096–105. doi:10.1128/jb.179.13.4096-4105.1997. PMC 179227 . PMID 9209021.