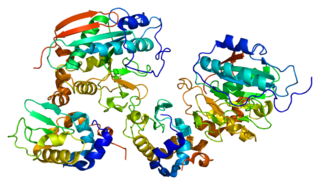Enzymes are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called enzymology and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties.
Tryptophan synthase or tryptophan synthetase is an enzyme that catalyzes the final two steps in the biosynthesis of tryptophan. It is commonly found in Eubacteria, Archaebacteria, Protista, Fungi, and Plantae. However, it is absent from Animalia. It is typically found as an α2β2 tetramer. The α subunits catalyze the reversible formation of indole and glyceraldehyde-3-phosphate (G3P) from indole-3-glycerol phosphate (IGP). The β subunits catalyze the irreversible condensation of indole and serine to form tryptophan in a pyridoxal phosphate (PLP) dependent reaction. Each α active site is connected to a β active site by a 25 Ångstrom long hydrophobic channel contained within the enzyme. This facilitates the diffusion of indole formed at α active sites directly to β active sites in a process known as substrate channeling. The active sites of tryptophan synthase are allosterically coupled.

Malate dehydrogenase (EC 1.1.1.37) (MDH) is an enzyme that reversibly catalyzes the oxidation of malate to oxaloacetate using the reduction of NAD+ to NADH. This reaction is part of many metabolic pathways, including the citric acid cycle. Other malate dehydrogenases, which have other EC numbers and catalyze other reactions oxidizing malate, have qualified names like malate dehydrogenase (NADP+).

Glycogen phosphorylase is one of the phosphorylase enzymes. Glycogen phosphorylase catalyzes the rate-limiting step in glycogenolysis in animals by releasing glucose-1-phosphate from the terminal alpha-1,4-glycosidic bond. Glycogen phosphorylase is also studied as a model protein regulated by both reversible phosphorylation and allosteric effects.
Glycogenin is an enzyme involved in converting glucose to glycogen. It acts as a primer, by polymerizing the first few glucose molecules, after which other enzymes take over. It is a homodimer of 37-kDa subunits and is classified as a glycosyltransferase.

1,4-alpha-glucan-branching enzyme, also known as brancher enzyme or glycogen-branching enzyme is an enzyme that in humans is encoded by the GBE1 gene.

The TIM barrel, also known as an alpha/beta barrel, is a conserved protein fold consisting of eight alpha helices (α-helices) and eight parallel beta strands (β-strands) that alternate along the peptide backbone. The structure is named after triose-phosphate isomerase, a conserved metabolic enzyme. TIM barrels are ubiquitous, with approximately 10% of all enzymes adopting this fold. Further, five of seven enzyme commission (EC) enzyme classes include TIM barrel proteins. The TIM barrel fold is evolutionarily ancient, with many of its members possessing little similarity today, instead falling within the twilight zone of sequence similarity.
α-Lactalbumin, also known as LALBA, is a protein that in humans is encoded by the LALBA gene.

α-Glucosidase (EC 3.2.1.20, is a glucosidase located in the brush border of the small intestine that acts upon α bonds:

Phosphorylase kinase (PhK) is a serine/threonine-specific protein kinase which activates glycogen phosphorylase to release glucose-1-phosphate from glycogen. PhK phosphorylates glycogen phosphorylase at two serine residues, triggering a conformational shift which favors the more active glycogen phosphorylase "a" form over the less active glycogen phosphorylase b.

Guanosine monophosphate synthetase, also known as GMPS is an enzyme that converts xanthosine monophosphate to guanosine monophosphate.
In enzymology, a cellobiose dehydrogenase (acceptor) (EC 1.1.99.18) is an enzyme that catalyzes the chemical reaction
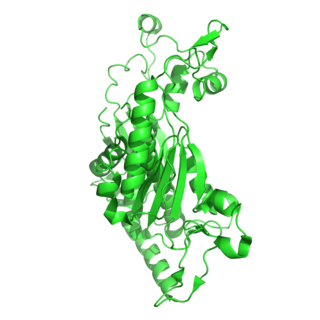
The enzyme chorismate synthase catalyzes the chemical reaction

Sucrose-phosphate synthase (SPS) is a plant enzyme involved in sucrose biosynthesis. Specifically, this enzyme catalyzes the transfer of a hexosyl group from uridine diphosphate glucose (UDP-glucose) to D-fructose 6-phosphate to form UDP and D-sucrose-6-phosphate. This reversible step acts as the key regulatory control point in sucrose biosynthesis, and is an excellent example of various key enzyme regulation strategies such as allosteric control and reversible phosphorylation.
In enzymology, a nucleoside-phosphate kinase is an enzyme that catalyzes the chemical reaction
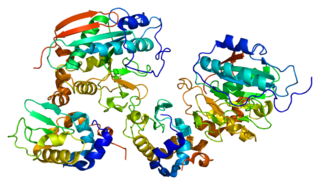
Beta-1,4-galactosyltransferase 1 is an enzyme that in humans is encoded by the B4GALT1 gene.

Beta-1,4-galactosyltransferase 2 is an enzyme that in humans is encoded by the B4GALT2 gene.

Lactose permease is a membrane protein which is a member of the major facilitator superfamily. Lactose permease can be classified as a symporter, which uses the proton gradient towards the cell to transport β-galactosides such as lactose in the same direction into the cell.
The Walker A and Walker B motifs are protein sequence motifs, known to have highly conserved three-dimensional structures. These were first reported in ATP-binding proteins by Walker and co-workers in 1982.
In molecular biology, glycoside hydrolase family 22 is a family of glycoside hydrolases. EC 3.2.1., which are a widespread group of enzymes that hydrolyse the glycosidic bond between two or more carbohydrates, or between a carbohydrate and a non-carbohydrate moiety. A classification system for glycoside hydrolases, based on sequence similarity, has led to the definition of >100 different families. This classification is available on the CAZy web site, and also discussed at CAZypedia, an online encyclopedia of carbohydrate active enzymes.