Related Research Articles

In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the most essential part of biological inheritance. This is essential for cell division during growth and repair of damaged tissues, while it also ensures that each of the new cells receives its own copy of the DNA. The cell possesses the distinctive property of division, which makes replication of DNA essential.
Mutagenesis is a process by which the genetic information of an organism is changed by the production of a mutation. It may occur spontaneously in nature, or as a result of exposure to mutagens. It can also be achieved experimentally using laboratory procedures. A mutagen is a mutation-causing agent, be it chemical or physical, which results in an increased rate of mutations in an organism's genetic code. In nature mutagenesis can lead to cancer and various heritable diseases, and it is also a driving force of evolution. Mutagenesis as a science was developed based on work done by Hermann Muller, Charlotte Auerbach and J. M. Robson in the first half of the 20th century.
In biochemistry, a polymerase is an enzyme that synthesizes long chains of polymers or nucleic acids. DNA polymerase and RNA polymerase are used to assemble DNA and RNA molecules, respectively, by copying a DNA template strand using base-pairing interactions or RNA by half ladder replication.

A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create two identical DNA duplexes from a single original DNA duplex. During this process, DNA polymerase "reads" the existing DNA strands to create two new strands that match the existing ones. These enzymes catalyze the chemical reaction
DNA primase is an enzyme involved in the replication of DNA and is a type of RNA polymerase. Primase catalyzes the synthesis of a short RNA segment called a primer complementary to a ssDNA template. After this elongation, the RNA piece is removed by a 5' to 3' exonuclease and refilled with DNA.

DNA polymerase I is an enzyme that participates in the process of prokaryotic DNA replication. Discovered by Arthur Kornberg in 1956, it was the first known DNA polymerase. It was initially characterized in E. coli and is ubiquitous in prokaryotes. In E. coli and many other bacteria, the gene that encodes Pol I is known as polA. The E. coli Pol I enzyme is composed of 928 amino acids, and is an example of a processive enzyme — it can sequentially catalyze multiple polymerisation steps without releasing the single-stranded template. The physiological function of Pol I is mainly to support repair of damaged DNA, but it also contributes to connecting Okazaki fragments by deleting RNA primers and replacing the ribonucleotides with DNA.

The SOS response is a global response to DNA damage in which the cell cycle is arrested and DNA repair and mutagenesis are induced. The system involves the RecA protein. The RecA protein, stimulated by single-stranded DNA, is involved in the inactivation of the repressor (LexA) of SOS response genes thereby inducing the response. It is an error-prone repair system that contributes significantly to DNA changes observed in a wide range of species.

DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA damage, resulting in tens of thousands of individual molecular lesions per cell per day. Many of these lesions cause structural damage to the DNA molecule and can alter or eliminate the cell's ability to transcribe the gene that the affected DNA encodes. Other lesions induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells after it undergoes mitosis. As a consequence, the DNA repair process is constantly active as it responds to damage in the DNA structure. When normal repair processes fail, and when cellular apoptosis does not occur, irreparable DNA damage may occur. This can eventually lead to malignant tumors, or cancer as per the two-hit hypothesis.

DNA polymerase II is a prokaryotic DNA-dependent DNA polymerase encoded by the PolB gene.
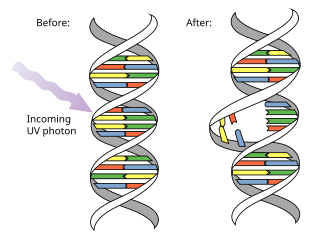
Pyrimidine dimers represent molecular lesions originating from thymine or cytosine bases within DNA, resulting from photochemical reactions. These lesions, commonly linked to direct DNA damage, are induced by ultraviolet light (UV), particularly UVC, result in the formation of covalent bonds between adjacent nitrogenous bases along the nucleotide chain near their carbon–carbon double bonds, the photo-coupled dimers are fluorescent. Such dimerization, which can also occur in double-stranded RNA (dsRNA) involving uracil or cytosine, leads to the creation of cyclobutane pyrimidine dimers (CPDs) and 6–4 photoproducts. These pre-mutagenic lesions modify the DNA helix structure, resulting in abnormal non-canonical base pairing and, consequently, adjacent thymines or cytosines in DNA will form a cyclobutane ring when joined together and cause a distortion in the DNA. This distortion prevents DNA replication and transcription mechanisms beyond the dimerization site.
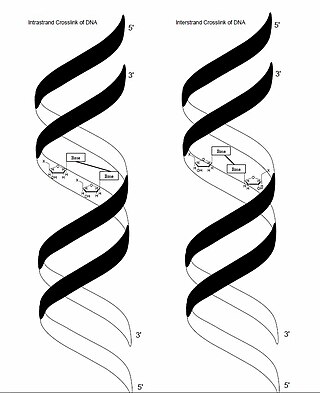
In genetics, crosslinking of DNA occurs when various exogenous or endogenous agents react with two nucleotides of DNA, forming a covalent linkage between them. This crosslink can occur within the same strand (intrastrand) or between opposite strands of double-stranded DNA (interstrand). These adducts interfere with cellular metabolism, such as DNA replication and transcription, triggering cell death. These crosslinks can, however, be repaired through excision or recombination pathways.
Postreplication repair is the repair of damage to the DNA that takes place after replication.
DNA polymerase kappa is a DNA polymerase that in humans is encoded by the POLK gene. It is involved in translesion synthesis.
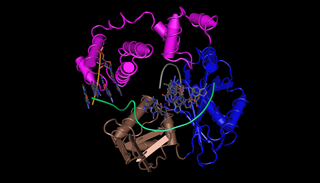
DNA polymerase lambda, also known as Pol λ, is an enzyme found in all eukaryotes. In humans, it is encoded by the POLL gene.

DNA repair protein REV1 is a protein that in humans is encoded by the REV1 gene.

Protein reversionless 3-like (REV3L) also known as DNA polymerase zeta catalytic subunit (POLZ) is an enzyme that in humans is encoded by the REV3L gene.

DNA polymerase eta, is a protein that in humans is encoded by the POLH gene.

In molecular biology, kataegis describes a pattern of localized hypermutations identified in some cancer genomes, in which a large number of highly patterned basepair mutations occur in a small region of DNA. The mutational clusters are usually several hundred basepairs long, alternating between a long range of C→T substitutional pattern and a long range of G→A substitutional pattern. This suggests that kataegis is carried out on only one of the two template strands of DNA during replication. Compared to other cancer-related mutations, such as chromothripsis, kataegis is more commonly seen; it is not an accumulative process but likely happens during one cycle of replication.
DNA polymerase IV is a prokaryotic polymerase that is involved in mutagenesis and is encoded by the dinB gene. It exhibits no 3′→5′ exonuclease (proofreading) activity and hence is error prone. In E. coli, DNA polymerase IV is involved in non-targeted mutagenesis. Pol IV is a Family Y polymerase expressed by the dinB gene that is switched on via SOS induction caused by stalled polymerases at the replication fork. During SOS induction, Pol IV production is increased tenfold and one of the functions during this time is to interfere with Pol III holoenzyme processivity. This creates a checkpoint, stops replication, and allows time to repair DNA lesions via the appropriate repair pathway. Another function of Pol IV is to perform translesion synthesis at the stalled replication fork like, for example, bypassing N2-deoxyguanine adducts at a faster rate than transversing undamaged DNA. Cells lacking dinB gene have a higher rate of mutagenesis caused by DNA damaging agents.

PrimPol is a protein encoded by the PRIMPOL gene in humans. PrimPol is a eukaryotic protein with both DNA polymerase and DNA Primase activities involved in translesion DNA synthesis. It is the first eukaryotic protein to be identified with priming activity using deoxyribonucleotides. It is also the first protein identified in the mitochondria to have translesion DNA synthesis activities.
References
- 1 2 3 Sutton MD, Walker GC (July 2001). "Managing DNA polymerases: coordinating DNA replication, DNA repair, and DNA recombination". Proceedings of the National Academy of Sciences of the United States of America. 98 (15): 8342–9. doi: 10.1073/pnas.111036998 . PMC 37441 . PMID 11459973.
- 1 2 Yang W (February 2003). "Damage repair DNA polymerases Y". Current Opinion in Structural Biology. 13 (1): 23–30. doi:10.1016/S0959-440X(02)00003-9. PMID 12581656.
- ↑ Garrett RH (2013). Biochemistry (1st Canadian ed.). Toronto: Nelson Education. p. 343. ISBN 9780176502652.
- 1 2 Goodman MF, Woodgate R (October 2013). "Translesion DNA polymerases". Cold Spring Harbor Perspectives in Biology. 5 (10): a010363. doi:10.1101/cshperspect.a010363. PMC 3783050 . PMID 23838442.
- 1 2 3 Jarosz DF, Beuning PJ, Cohen SE, Walker GC (February 2007). "Y-family DNA polymerases in Escherichia coli". Trends in Microbiology. 15 (2): 70–7. doi:10.1016/j.tim.2006.12.004. hdl: 1721.1/70041 . PMID 17207624.
- 1 2 Patel M, Jiang Q, Woodgate R, Cox MM, Goodman MF (June 2010). "A new model for SOS-induced mutagenesis: how RecA protein activates DNA polymerase V". Critical Reviews in Biochemistry and Molecular Biology. 45 (3): 171–84. doi:10.3109/10409238.2010.480968. PMC 2874081 . PMID 20441441.
- 1 2 Yang W (May 2014). "An overview of Y-Family DNA polymerases and a case study of human DNA polymerase η". Biochemistry. 53 (17): 2793–803. doi:10.1021/bi500019s. PMC 4018060 . PMID 24716551.
- 1 2 3 Fuchs RP, Fujii S (December 2013). "Translesion DNA synthesis and mutagenesis in prokaryotes". Cold Spring Harbor Perspectives in Biology. 5 (12): a012682. doi:10.1101/cshperspect.a012682. PMC 3839610 . PMID 24296168.
- ↑ Doles J, Oliver TG, Cameron ER, Hsu G, Jacks T, Walker GC, Hemann MT (November 2010). "Suppression of Rev3, the catalytic subunit of Pol{zeta}, sensitizes drug-resistant lung tumors to chemotherapy". Proceedings of the National Academy of Sciences of the United States of America. 107 (48): 20786–91. doi: 10.1073/pnas.1011409107 . PMC 2996428 . PMID 21068376.