| DNA pol III theta subunit | |||||||
|---|---|---|---|---|---|---|---|
| Identifiers | |||||||
| Organism | |||||||
| Symbol | holE | ||||||
| Entrez | 947471 | ||||||
| RefSeq (Prot) | NP_416356.1 | ||||||
| UniProt | P0ABS8 | ||||||
| Other data | |||||||
| EC number | 2.7.7.7 | ||||||
| Chromosome | genome: 1.92 - 1.92 Mb | ||||||
| |||||||
| DNA polymerase III, theta subunit | |||||||||
|---|---|---|---|---|---|---|---|---|---|
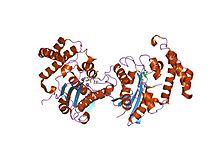 Structure of the E. coli Pol III epsilon-Hot proofreading complex | |||||||||
| Identifiers | |||||||||
| Symbol | DNA_pol3_theta | ||||||||
| Pfam | PF06440 | ||||||||
| InterPro | IPR009052 | ||||||||
| SCOP2 | 1du2 / SCOPe / SUPFAM | ||||||||
| |||||||||
In E. coli and other bacteria, holE is a gene that encodes the theta subunit of DNA polymerase III. [1]