
In molecular biology, RNA polymerase, is an enzyme that synthesizes RNA from a DNA template.

A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create two identical DNA duplexes from a single original DNA duplex. During this process, DNA polymerase "reads" the existing DNA strands to create two new strands that match the existing ones. These enzymes catalyze the chemical reaction

DNA polymerase III holoenzyme is the primary enzyme complex involved in prokaryotic DNA replication. It was discovered by Thomas Kornberg and Malcolm Gefter in 1970. The complex has high processivity and, specifically referring to the replication of the E.coli genome, works in conjunction with four other DNA polymerases. Being the primary holoenzyme involved in replication activity, the DNA Pol III holoenzyme also has proofreading capabilities that corrects replication mistakes by means of exonuclease activity reading 3'→5' and synthesizing 5'→3'. DNA Pol III is a component of the replisome, which is located at the replication fork.
dnaQ is the gene encoding the ε subunit of DNA polymerase III in Escherichia coli. The ε subunit is one of three core proteins in the DNA polymerase complex. It functions as a 3’→5’ DNA directed proofreading exonuclease that removes incorrectly incorporated bases during replication. dnaQ may also be referred to as mutD.
In molecular biology and biochemistry, processivity is an enzyme's ability to catalyze "consecutive reactions without releasing its substrate".
DNA gyrase, or simply gyrase, is an enzyme within the class of topoisomerase and is a subclass of Type II topoisomerases that reduces topological strain in an ATP dependent manner while double-stranded DNA is being unwound by elongating RNA-polymerase or by helicase in front of the progressing replication fork. The enzyme causes negative supercoiling of the DNA or relaxes positive supercoils. It does so by looping the template so as to form a crossing, then cutting one of the double helices and passing the other through it before releasing the break, changing the linking number by two in each enzymatic step. This process occurs in bacteria, whose single circular DNA is cut by DNA gyrase and the two ends are then twisted around each other to form supercoils. Gyrase is also found in eukaryotic plastids: it has been found in the apicoplast of the malarial parasite Plasmodium falciparum and in chloroplasts of several plants. Bacterial DNA gyrase is the target of many antibiotics, including nalidixic acid, novobiocin, and ciprofloxacin.

Exonucleases are enzymes that work by cleaving nucleotides one at a time from the end (exo) of a polynucleotide chain. A hydrolyzing reaction that breaks phosphodiester bonds at either the 3′ or the 5′ end occurs. Its close relative is the endonuclease, which cleaves phosphodiester bonds in the middle (endo) of a polynucleotide chain. Eukaryotes and prokaryotes have three types of exonucleases involved in the normal turnover of mRNA: 5′ to 3′ exonuclease (Xrn1), which is a dependent decapping protein; 3′ to 5′ exonuclease, an independent protein; and poly(A)-specific 3′ to 5′ exonuclease.

DNA polymerase II is a prokaryotic DNA-Dependent DNA polymerase encoded by the PolB gene.
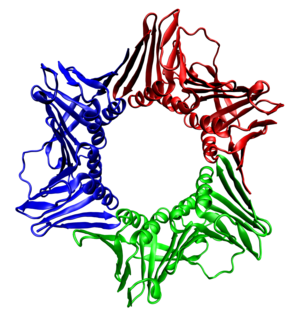
Proliferating cell nuclear antigen (PCNA) is a DNA clamp that acts as a processivity factor for DNA polymerase δ in eukaryotic cells and is essential for replication. PCNA is a homotrimer and achieves its processivity by encircling the DNA, where it acts as a scaffold to recruit proteins involved in DNA replication, DNA repair, chromatin remodeling and epigenetics.

A DNA clamp, also known as a sliding clamp or β-clamp, is a protein complex that serves as a processivity-promoting factor in DNA replication. As a critical component of the DNA polymerase III holoenzyme, the clamp protein binds DNA polymerase and prevents this enzyme from dissociating from the template DNA strand. The clamp-polymerase protein–protein interactions are stronger and more specific than the direct interactions between the polymerase and the template DNA strand; because one of the rate-limiting steps in the DNA synthesis reaction is the association of the polymerase with the DNA template, the presence of the sliding clamp dramatically increases the number of nucleotides that the polymerase can add to the growing strand per association event. The presence of the DNA clamp can increase the rate of DNA synthesis up to 1,000-fold compared with a nonprocessive polymerase.
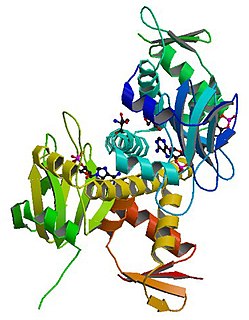
cAMP receptor protein is a regulatory protein in bacteria. CRP protein binds cAMP, which causes a conformational change that allows CRP to bind tightly to a specific DNA site in the promoters of the genes it controls. CRP then activates transcription through direct protein–protein interactions with RNA polymerase.
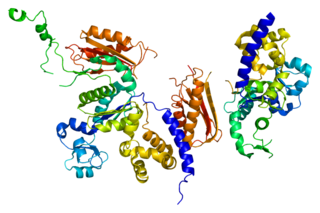
DNA polymerase kappa is an DNA polymerase that in humans is encoded by the POLK gene. It is involved in translesion synthesis.

In molecular biology, the protein domain SAICAR synthase is an enzyme which catalyses a reaction to create SAICAR. In enzymology, this enzyme is also known as phosphoribosylaminoimidazolesuccinocarboxamide synthase. It is an enzyme that catalyzes the chemical reaction

In enzymology, a dUTP diphosphatase (EC 3.6.1.23) is an enzyme that catalyzes the chemical reaction

Endonuclease III-like protein 1 is an enzyme that in humans is encoded by the NTHL1 gene.

A circular chromosome is a chromosome in bacteria, archaea, mitochondria, and chloroplasts, in the form of a molecule of circular DNA, unlike the linear chromosome of most eukaryotes.
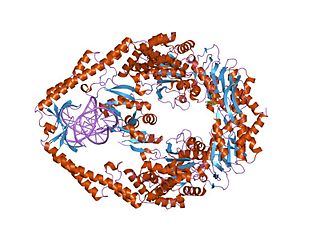
MutS is a mismatch DNA repair protein, originally described in Escherichia coli.

In molecular biology, the ars operon is an operon found in several bacterial taxon. It is required for the detoxification of arsenate, arsenite, and antimonite. This system transports arsenite and antimonite out of the cell. The pump is composed of two polypeptides, the products of the arsA and arsB genes. This two-subunit enzyme produces resistance to arsenite and antimonite. Arsenate, however, must first be reduced to arsenite before it is extruded. A third gene, arsC, expands the substrate specificity to allow for arsenate pumping and resistance. ArsC is an approximately 150-residue arsenate reductase that uses reduced glutathione (GSH) to convert arsenate to arsenite with a redox active cysteine residue in the active site. ArsC forms an active quaternary complex with GSH, arsenate, and glutaredoxin 1 (Grx1). The three ligands must be present simultaneously for reduction to occur.

In molecular biology, the δ (delta) subunit of DNA polymerase III is encoded by the holA gene in E. coli and other bacteria. Along with the γ, δ', χ, and ψ subunits that make up the core polymerase, and the β accessory proteins, the δ subunit is responsible for the high speed and processivity of polIII.
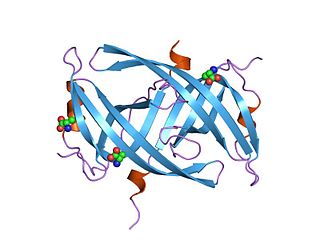
Single-stranded binding proteins (SSBs) are a class of proteins that have been identified in both viruses and organisms from bacteria to humans.

















