
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the most essential part for biological inheritance. This is essential for cell division during growth and repair of damaged tissues, while it also ensures that each of the new cells receives its own copy of the DNA. The cell possesses the distinctive property of division, which makes replication of DNA essential.

Transcription is the process of copying a segment of DNA into RNA. The segments of DNA transcribed into RNA molecules that can encode proteins are said to produce messenger RNA (mRNA). Other segments of DNA are copied into RNA molecules called non-coding RNAs (ncRNAs). mRNA comprises only 1–3% of total RNA samples. Less than 2% of the human genome can be transcribed into mRNA, while at least 80% of mammalian genomic DNA can be actively transcribed, with the majority of this 80% considered to be ncRNA.

In molecular biology, RNA polymerase, or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that synthesizes RNA from a DNA template.

A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create two identical DNA duplexes from a single original DNA duplex. During this process, DNA polymerase "reads" the existing DNA strands to create two new strands that match the existing ones. These enzymes catalyze the chemical reaction
DNA primase is an enzyme involved in the replication of DNA and is a type of RNA polymerase. Primase catalyzes the synthesis of a short RNA segment called a primer complementary to a ssDNA template. After this elongation, the RNA piece is removed by a 5' to 3' exonuclease and refilled with DNA.

DNA polymerase III holoenzyme is the primary enzyme complex involved in prokaryotic DNA replication. It was discovered by Thomas Kornberg and Malcolm Gefter in 1970. The complex has high processivity and, specifically referring to the replication of the E.coli genome, works in conjunction with four other DNA polymerases. Being the primary holoenzyme involved in replication activity, the DNA Pol III holoenzyme also has proofreading capabilities that corrects replication mistakes by means of exonuclease activity reading 3'→5' and synthesizing 5'→3'. DNA Pol III is a component of the replisome, which is located at the replication fork.
In molecular biology and biochemistry, processivity is an enzyme's ability to catalyze "consecutive reactions without releasing its substrate".

Okazaki fragments are short sequences of DNA nucleotides which are synthesized discontinuously and later linked together by the enzyme DNA ligase to create the lagging strand during DNA replication. They were discovered in the 1960s by the Japanese molecular biologists Reiji and Tsuneko Okazaki, along with the help of some of their colleagues.

General transcription factors (GTFs), also known as basal transcriptional factors, are a class of protein transcription factors that bind to specific sites (promoter) on DNA to activate transcription of genetic information from DNA to messenger RNA. GTFs, RNA polymerase, and the mediator constitute the basic transcriptional apparatus that first bind to the promoter, then start transcription. GTFs are also intimately involved in the process of gene regulation, and most are required for life.
The replication factor C, or RFC, is a five-subunit protein complex that is required for DNA replication.

The replisome is a complex molecular machine that carries out replication of DNA. The replisome first unwinds double stranded DNA into two single strands. For each of the resulting single strands, a new complementary sequence of DNA is synthesized. The Total result is formation of two new double stranded DNA sequences that are exact copies of the original double stranded DNA sequence.

Proliferating cell nuclear antigen (PCNA) is a DNA clamp that acts as a processivity factor for DNA polymerase δ in eukaryotic cells and is essential for replication. PCNA is a homotrimer and achieves its processivity by encircling the DNA, where it acts as a scaffold to recruit proteins involved in DNA replication, DNA repair, chromatin remodeling and epigenetics.

Eukaryotic DNA replication is a conserved mechanism that restricts DNA replication to once per cell cycle. Eukaryotic DNA replication of chromosomal DNA is central for the duplication of a cell and is necessary for the maintenance of the eukaryotic genome.

Bacterial transcription is the process in which a segment of bacterial DNA is copied into a newly synthesized strand of messenger RNA (mRNA) with use of the enzyme RNA polymerase.

Replication factor C subunit 4 is a protein that in humans is encoded by the RFC4 gene.

Replication factor C subunit 5 is a protein that in humans is encoded by the RFC5 gene.
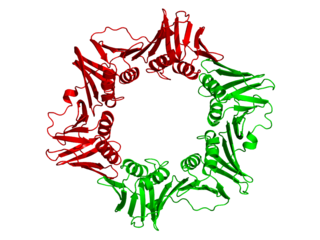
dnaN is the gene that codes for the DNA clamp of DNA polymerase III in prokaryotes. The β clamp physically locks Pol III onto a DNA strand during replication to help increase its processivity. The eukaryotic equivalent to the β clamp is PCNA.
DNA polymerase delta(DNA Pol δ) is an enzyme complex found in eukaryotes that is involved in DNA replication and repair. The DNA polymerase delta complex consists of 4 subunits: POLD1, POLD2, POLD3, and POLD4. DNA Pol δ is an enzyme used for both leading and lagging strand synthesis. It exhibits increased processivity when interacting with the proliferating cell nuclear antigen (PCNA). As well, the multisubunit protein replication factor C, through its role as the clamp loader for PCNA is important for DNA Pol δ function.
DNA polymerase epsilon is a member of the DNA polymerase family of enzymes found in eukaryotes. It is composed of the following four subunits: POLE, POLE2, POLE3, and POLE4. Recent evidence suggests that it plays a major role in leading strand DNA synthesis and nucleotide and base excision repair.

Michael E. O’ Donnell is an American biochemist and a professor at the Rockefeller University specializing in the field of DNA replication.





















