
Sequence homology is the biological homology between DNA, RNA, or protein sequences, defined in terms of shared ancestry in the evolutionary history of life. Two segments of DNA can have shared ancestry because of three phenomena: either a speciation event (orthologs), or a duplication event (paralogs), or else a horizontal gene transfer event (xenologs).

Pfam is a database of protein families that includes their annotations and multiple sequence alignments generated using hidden Markov models. The most recent version, Pfam 33.1, was released in May 2020 and contains 18,259 families.

Amos Bairoch is a Swiss bioinformatician and Professor of Bioinformatics at the Department of Human Protein Sciences of the University of Geneva where he leads the CALIPHO group at the Swiss Institute of Bioinformatics (SIB) combining bioinformatics, curation, and experimental efforts to functionally characterize human proteins.
InterPro is a database of protein families, domains and functional sites in which identifiable features found in known proteins can be applied to new protein sequences in order to functionally characterise them.

A protein domain is a conserved part of a given protein sequence and tertiary structure that can evolve, function, and exist independently of the rest of the protein chain. Each domain forms a compact three-dimensional structure and often can be independently stable and folded. Many proteins consist of several structural domains. One domain may appear in a variety of different proteins. Molecular evolution uses domains as building blocks and these may be recombined in different arrangements to create proteins with different functions. In general, domains vary in length from between about 50 amino acids up to 250 amino acids in length. The shortest domains, such as zinc fingers, are stabilized by metal ions or disulfide bridges. Domains often form functional units, such as the calcium-binding EF hand domain of calmodulin. Because they are independently stable, domains can be "swapped" by genetic engineering between one protein and another to make chimeric proteins.

MicrobesOnline is a publicly and freely accessible website that hosts multiple comparative genomic tools for comparing microbial species at the genomic, transcriptomic and functional levels. MicrobesOnline was developed by the Virtual Institute for Microbial Stress and Survival, which is based at the Lawrence Berkeley National Laboratory in Berkeley, California. The site was launched in 2005, with regular updates until 2011.
The Eukaryotic Linear Motif (ELM) resource is a computational biology resource for investigating short linear motifs (SLiMs) in eukaryotic proteins. It is currently the largest collection of linear motif classes with annotated and experimentally validated linear motif instances.

Christopher Paul Ponting is a British computational biologist, specializing in the evolution and function of genes and genomes. He is currently Chair of Medical Bioinformatics at the University of Edinburgh and group leader in the MRC Human Genetics Unit. He is also an Associate Faculty member of the Wellcome Trust Sanger Institute, a Fellow of the Academy of Medical Sciences, member of the European Molecular Biology Organisation and Fellow of the Royal Society of Edinburgh. His research focuses on long noncoding RNA function and evolution, on single cell biology and on disease genomics.

In molecular biology, STRING is a biological database and web resource of known and predicted protein–protein interactions.
SUPERFAMILY is a database and search platform of structural and functional annotation for all proteins and genomes. It classifies amino acid sequences into known structural domains, especially into SCOP superfamilies. Domains are functional, structural, and evolutionary units that form proteins. Domains of common Ancestry are grouped into superfamilies. The domains and domain superfamilies are defined and described in SCOP. Superfamilies are groups of proteins which have structural evidence to support a common evolutionary ancestor but may not have detectable sequence homology.
A domain of unknown function (DUF) is a protein domain that has no characterised function. These families have been collected together in the Pfam database using the prefix DUF followed by a number, with examples being DUF2992 and DUF1220. As of 2019, there are almost 4,000 DUF families within the Pfam database representing over 22% of known families. Some DUFs are not named using the nomenclature due to popular usage but are nevertheless DUFs.
PDBsum is a database that provides an overview of the contents of each 3D macromolecular structure deposited in the Protein Data Bank. The original version of the database was developed around 1995 by Roman Laskowski and collaborators at University College London. As of 2014, PDBsum is maintained by Laskowski and collaborators in the laboratory of Janet Thornton at the European Bioinformatics Institute (EBI).
The eggNOG database is a database of biological information hosted by the EMBL. It is based on the original idea of COGs and expands that idea to non-supervised orthologous groups constructed from numerous organisms. The database was created in 2007 and updated to version 4.5 in 2015. eggNOG stands for evolutionary genealogy of genes: Non-supervised Orthologous Groups.
αr9 is a family of bacterial small non-coding RNAs with representatives in a broad group of α-proteobacteria from the order Rhizobiales. The first member of this family (Smr9C) was found in a Sinorhizobium meliloti 1021 locus located in the chromosome (C). Further homology and structure conservation analysis have identified full-length Smr9C homologs in several nitrogen-fixing symbiotic rhizobia, in the plant pathogens belonging to Agrobacterium species as well as in a broad spectrum of Brucella species. αr9C RNA species are 144-158 nt long and share a well defined common secondary structure consisting of seven conserved regions. Most of the αr9 transcripts can be catalogued as trans-acting sRNAs expressed from well-defined promoter regions of independent transcription units within intergenic regions (IGRs) of the α-proteobacterial genomes.
TIGRFAMs is a database of protein families designed to support manual and automated genome annotation. Each entry includes a multiple sequence alignment and hidden Markov model (HMM) built from the alignment. Sequences that score above the defined cutoffs of a given TIGRFAMs HMM are assigned to that protein family and may be assigned the corresponding annotations.
Megf8 also known as Multiple Epidermal Growth Factor-like Domains 8, is a protein coding gene that encodes a single pass membrane protein, known to participate in developmental regulation and cellular communication. It is located on chromosome 19 at the 49th open reading frame in humans (19q13.2). There are two isoform constructs known for MEGF8, which differ by a 67 amino acid indel. The isoform 2 splice version is 2785 amino acids long, and predicted to be 296.6 kdal in mass. Isoform 1 is composed of 2845 amino acids and predicted to weigh 303.1 kdal. Using BLAST searches, orthologs were found primarily in mammals, but MEGF8 is also conserved in invertebrates and fishes, and rarely in birds, reptiles, and amphibians. A notably important paralog to multiple epidermal growth factor-like domains 8 is ATRNL1, which is also a single pass transmembrane protein, with several of the same key features and motifs as MEGF8, as indicated by Simple Modular Architecture Research Tool (SMART) which is hosted by the European Molecular Biology Laboratory located in Heidelberg, Germany. MEGF8 has been predicted to be a key player in several developmental processes, such as left-right patterning and limb formation. Currently, researchers have found MEGF8 SNP mutations to be the cause of Carpenter syndrome subtype 2.
Julian John Thurstan Gough is a Group Leader in the Laboratory of Molecular Biology (LMB) of the Medical Research Council (MRC). He was previously a professor of Bioinformatics at the University of Bristol.

Christine Anne Orengo is a Professor of Bioinformatics at University College London (UCL) known for her work on protein structure, particularly the CATH database. Orengo serves as president of the International Society for Computational Biology (ISCB), the first woman to do so in the history of the society.
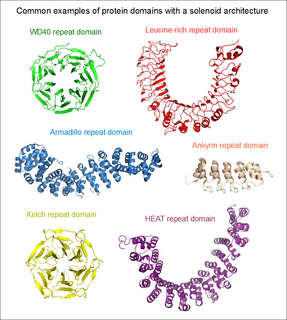
An array of protein tandem repeats is defined as several adjacent copies having the same or similar sequence motifs. These periodic sequences are generated by internal duplications in both coding and non-coding genomic sequences. Repetitive units of protein tandem repeats are considerably diverse, ranging from the repetition of a single amino acid to domains of 100 or more residues.









