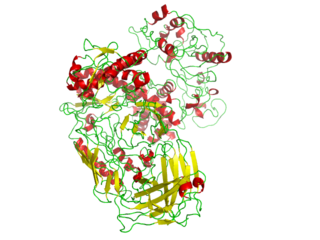
In biochemistry, a polymerase is an enzyme that synthesizes long chains of polymers or nucleic acids. DNA polymerase and RNA polymerase are used to assemble DNA and RNA molecules, respectively, by copying a DNA template strand using base-pairing interactions or RNA by half ladder replication.

Transcription is the process of copying a segment of DNA into RNA. The segments of DNA transcribed into RNA molecules that can encode proteins produce messenger RNA (mRNA). Other segments of DNA are transcribed into RNA molecules called non-coding RNAs (ncRNAs).

In molecular biology, RNA polymerase, or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that catalyzes the chemical reactions that synthesize RNA from a DNA template.
A sigma factor is a protein needed for initiation of transcription in bacteria. It is a bacterial transcription initiation factor that enables specific binding of RNA polymerase (RNAP) to gene promoters. It is homologous to archaeal transcription factor B and to eukaryotic factor TFIIB. The specific sigma factor used to initiate transcription of a given gene will vary, depending on the gene and on the environmental signals needed to initiate transcription of that gene. Selection of promoters by RNA polymerase is dependent on the sigma factor that associates with it. They are also found in plant chloroplasts as a part of the bacteria-like plastid-encoded polymerase (PEP).

The preinitiation complex is a complex of approximately 100 proteins that is necessary for the transcription of protein-coding genes in eukaryotes and archaea. The preinitiation complex positions RNA polymerase II at gene transcription start sites, denatures the DNA, and positions the DNA in the RNA polymerase II active site for transcription.

RNA polymerase II is a multiprotein complex that transcribes DNA into precursors of messenger RNA (mRNA) and most small nuclear RNA (snRNA) and microRNA. It is one of the three RNAP enzymes found in the nucleus of eukaryotic cells. A 550 kDa complex of 12 subunits, RNAP II is the most studied type of RNA polymerase. A wide range of transcription factors are required for it to bind to upstream gene promoters and begin transcription.

General transcription factors (GTFs), also known as basal transcriptional factors, are a class of protein transcription factors that bind to specific sites (promoter) on DNA to activate transcription of genetic information from DNA to messenger RNA. GTFs, RNA polymerase, and the mediator constitute the basic transcriptional apparatus that first bind to the promoter, then start transcription. GTFs are also intimately involved in the process of gene regulation, and most are required for life.

Catabolite activator protein is a trans-acting transcriptional activator that exists as a homodimer in solution. Each subunit of CAP is composed of a ligand-binding domain at the N-terminus and a DNA-binding domain at the C-terminus. Two cAMP molecules bind dimeric CAP with negative cooperativity. Cyclic AMP functions as an allosteric effector by increasing CAP's affinity for DNA. CAP binds a DNA region upstream from the DNA binding site of RNA Polymerase. CAP activates transcription through protein-protein interactions with the α-subunit of RNA Polymerase. This protein-protein interaction is responsible for (i) catalyzing the formation of the RNAP-promoter closed complex; and (ii) isomerization of the RNAP-promoter complex to the open conformation. CAP's interaction with RNA polymerase causes bending of the DNA near the transcription start site, thus effectively catalyzing the transcription initiation process. CAP's name is derived from its ability to affect transcription of genes involved in many catabolic pathways. For example, when the amount of glucose transported into the cell is low, a cascade of events results in the increase of cytosolic cAMP levels. This increase in cAMP levels is sensed by CAP, which goes on to activate the transcription of many other catabolic genes.
In eukaryote cells, RNA polymerase III is a protein that transcribes DNA to synthesize 5S ribosomal RNA, tRNA, and other small RNAs.

A transcription bubble is a molecular structure formed during DNA transcription when a limited portion of the DNA double helix is unwound. The size of a transcription bubble ranges from 12 to 14 base pairs. A transcription bubble is formed when the RNA polymerase enzyme binds to a promoter and causes two DNA strands to detach. It presents a region of unpaired DNA, where a short stretch of nucleotides are exposed on each strand of the double helix.
Antitermination is the prokaryotic cell's aid to fix premature termination of RNA synthesis during the transcription of RNA. It occurs when the RNA polymerase ignores the termination signal and continues elongating its transcript until a second signal is reached. Antitermination provides a mechanism whereby one or more genes at the end of an operon can be switched either on or off, depending on the polymerase either recognizing or not recognizing the termination signal.

Bacterial transcription is the process in which a segment of bacterial DNA is copied into a newly synthesized strand of messenger RNA (mRNA) with use of the enzyme RNA polymerase.

Eukaryotic transcription is the elaborate process that eukaryotic cells use to copy genetic information stored in DNA into units of transportable complementary RNA replica. Gene transcription occurs in both eukaryotic and prokaryotic cells. Unlike prokaryotic RNA polymerase that initiates the transcription of all different types of RNA, RNA polymerase in eukaryotes comes in three variations, each translating a different type of gene. A eukaryotic cell has a nucleus that separates the processes of transcription and translation. Eukaryotic transcription occurs within the nucleus where DNA is packaged into nucleosomes and higher order chromatin structures. The complexity of the eukaryotic genome necessitates a great variety and complexity of gene expression control.

Intrinsic, or rho-independent termination, is a process to signal the end of transcription and release the newly constructed RNA molecule. In bacteria such as E. coli, transcription is terminated either by a rho-dependent process or rho-independent process. In the Rho-dependent process, the rho-protein locates and binds the signal sequence in the mRNA and signals for cleavage. Contrarily, intrinsic termination does not require a special protein to signal for termination and is controlled by the specific sequences of RNA. When the termination process begins, the transcribed mRNA forms a stable secondary structure hairpin loop, also known as a stem-loop. This RNA hairpin is followed by multiple uracil nucleotides. The bonds between uracil (rU) and adenine (dA) are very weak. A protein bound to RNA polymerase (nusA) binds to the stem-loop structure tightly enough to cause the polymerase to temporarily stall. This pausing of the polymerase coincides with transcription of the poly-uracil sequence. The weak adenine-uracil bonds lower the energy of destabilization for the RNA-DNA duplex, allowing it to unwind and dissociate from the RNA polymerase. Overall, the modified RNA structure is what terminates transcription.

DNA-directed RNA polymerase, mitochondrial is an enzyme that in humans is encoded by the POLRMT gene.
RNA polymerase II holoenzyme is a form of eukaryotic RNA polymerase II that is recruited to the promoters of protein-coding genes in living cells. It consists of RNA polymerase II, a subset of general transcription factors, and regulatory proteins known as SRB proteins.

DSIF is a protein complex that can either negatively or positively affect transcription by RNA polymerase II. It can interact with the negative elongation factor (NELF) to promote the stalling of Pol II at some genes, which is called promoter proximal pausing. The pause occurs soon after initiation, once 20-60 nucleotides have been transcribed. This stalling is relieved by positive transcription elongation factor b (P-TEFb) and Pol II enters productive elongation to resume synthesis till finish. In humans, DSIF is composed of hSPT4 and hSPT5. hSPT5 has a direct role in mRNA capping which occurs while the elongation is paused.
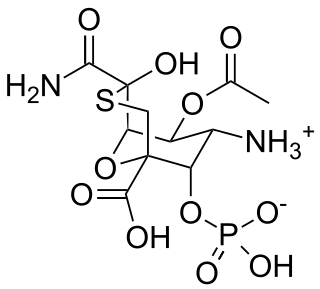
Tagetitoxin (TGT) is a bacterial phytotoxin produced by Pseudomonas syringae pv. tagetis.

Archaeal transcription factor B is a protein family of extrinsic transcription factors that guide the initiation of RNA transcription in organisms that fall under the domain of Archaea. It is homologous to eukaryotic TFIIB and, more distantly, to bacterial sigma factor. Like these proteins, it is involved in forming transcription preinitiation complexes. Its structure includes several conserved motifs which interact with DNA and other transcription factors, notably the single type of RNA polymerase that performs transcription in Archaea.
Phage-assisted continuous evolution (PACE) is a phage-based technique for the automated directed evolution of proteins. It relies on relating the desired activity of a target protein with the fitness of an infectious bacteriophage which carries the protein's corresponding gene. Proteins with greater desired activity hence confer greater infectivity to their carrier phage. More infectious phage propagate more effectively, selecting for advantageous mutations. Genetic variation is generated using error-prone polymerases on the phage vectors, and over time the protein accumulates beneficial mutations. This technique is notable for performing hundreds of rounds of selection with minimal human intervention.













