
Molecular dynamics (MD) is a computer simulation method for analyzing the physical movements of atoms and molecules. The atoms and molecules are allowed to interact for a fixed period of time, giving a view of the dynamic "evolution" of the system. In the most common version, the trajectories of atoms and molecules are determined by numerically solving Newton's equations of motion for a system of interacting particles, where forces between the particles and their potential energies are often calculated using interatomic potentials or molecular mechanical force fields. The method is applied mostly in chemical physics, materials science, and biophysics.

James Nicholas Gray was an American computer scientist who received the Turing Award in 1998 "for seminal contributions to database and transaction processing research and technical leadership in system implementation".
Folding@home is a distributed computing project aimed to help scientists develop new therapeutics for a variety of diseases by the means of simulating protein dynamics. This includes the process of protein folding and the movements of proteins, and is reliant on simulations run on volunteers' personal computers. Folding@home is currently based at the University of Pennsylvania and led by Greg Bowman, a former student of Vijay Pande.

Crowd simulation is the process of simulating the movement of a large number of entities or characters. It is commonly used to create virtual scenes for visual media like films and video games, and is also used in crisis training, architecture and urban planning, and evacuation simulation.
Molecular graphics is the discipline and philosophy of studying molecules and their properties through graphical representation. IUPAC limits the definition to representations on a "graphical display device". Ever since Dalton's atoms and Kekulé's benzene, there has been a rich history of hand-drawn atoms and molecules, and these representations have had an important influence on modern molecular graphics.

Fluid animation refers to computer graphics techniques for generating realistic animations of fluids such as water and smoke. Fluid animations are typically focused on emulating the qualitative visual behavior of a fluid, with less emphasis placed on rigorously correct physical results, although they often still rely on approximate solutions to the Euler equations or Navier–Stokes equations that govern real fluid physics. Fluid animation can be performed with different levels of complexity, ranging from time-consuming, high-quality animations for films, or visual effects, to simple and fast animations for real-time animations like computer games.
A massively parallel processor array, also known as a multi purpose processor array (MPPA) is a type of integrated circuit which has a massively parallel array of hundreds or thousands of CPUs and RAM memories. These processors pass work to one another through a reconfigurable interconnect of channels. By harnessing a large number of processors working in parallel, an MPPA chip can accomplish more demanding tasks than conventional chips. MPPAs are based on a software parallel programming model for developing high-performance embedded system applications.

Anton is a massively parallel supercomputer designed and built by D. E. Shaw Research in New York, first running in 2008. It is a special-purpose system for molecular dynamics (MD) simulations of proteins and other biological macromolecules. An Anton machine consists of a substantial number of application-specific integrated circuits (ASICs), interconnected by a specialized high-speed, three-dimensional torus network.
Desmond is a software package developed at D. E. Shaw Research to perform high-speed molecular dynamics simulations of biological systems on conventional computer clusters. The code uses novel parallel algorithms and numerical methods to achieve high performance on platforms containing multiple processors, but may also be executed on a single computer.
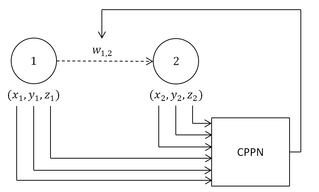
Hypercube-based NEAT, or HyperNEAT, is a generative encoding that evolves artificial neural networks (ANNs) with the principles of the widely used NeuroEvolution of Augmented Topologies (NEAT) algorithm developed by Kenneth Stanley. It is a novel technique for evolving large-scale neural networks using the geometric regularities of the task domain. It uses Compositional Pattern Producing Networks (CPPNs), which are used to generate the images for Picbreeder.orgArchived 2011-07-25 at the Wayback Machine and shapes for EndlessForms.comArchived 2018-11-14 at the Wayback Machine. HyperNEAT has recently been extended to also evolve plastic ANNs and to evolve the location of every neuron in the network.
Adaptive sampling is a technique used in computational molecular biology to efficiently simulate protein folding when coupled with molecular dynamics simulations.
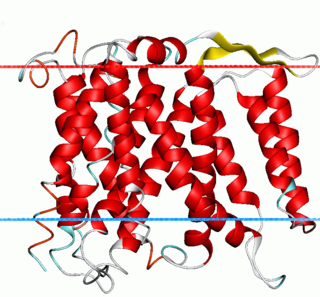
Na+/H+ antiporter A (NhaA) family (TC# 2.A.33) contains a number of bacterial sodium-proton antiporter (SPAP) proteins. These are integral membrane proteins that catalyse the exchange of H+ for Na+ in a manner that is highly pH dependent. Homologues have been sequenced from a number of bacteria and archaea. Prokaryotes possess multiple paralogues. A representative list of the proteins that belong to the NhaA family can be found in the Transporter Classification Database.
David Bacon is an American computer programmer.
Edmond Chow is a full professor in the School of Computational Science and Engineering of Georgia Institute of Technology. His main areas of research are in designing numerical methods for high-performance computing and applying these methods to solve large-scale scientific computing problems.
Thomas K. Porter is the senior vice president of production strategy at Pixar and one of the studio's founding employees.
Daniel Ramot is Israeli-born American entrepreneur, and scientist. He is the co-founder and CEO of TransitTech company, Via.
Andrew Thomas Campbell is a computer scientist who works in the field of ubiquitous computing. He is best known for his research on mobile sensing, applied machine learning and human behavioral modeling.
A domain-specific architecture (DSA) is a programmable computer architecture specifically tailored to operate very efficiently within the confines of a given application domain. The term is often used in contrast to general-purpose architectures, such as CPUs, that are designed to operate on any computer program.







