Related Research Articles

Computational chemistry is a branch of chemistry that uses computer simulations to assist in solving chemical problems. It uses methods of theoretical chemistry incorporated into computer programs to calculate the structures and properties of molecules, groups of molecules, and solids. The importance of this subject stems from the fact that, with the exception of some relatively recent findings related to the hydrogen molecular ion, achieving an accurate quantum mechanical depiction of chemical systems analytically, or in a closed form, is not feasible. The complexity inherent in the many-body problem exacerbates the challenge of providing detailed descriptions of quantum mechanical systems. While computational results normally complement information obtained by chemical experiments, it can occasionally predict unobserved chemical phenomena.
NWChem is an ab initio computational chemistry software package which includes quantum chemical and molecular dynamics functionality. It was designed to run on high-performance parallel supercomputers as well as conventional workstation clusters. It aims to be scalable both in its ability to treat large problems efficiently, and in its usage of available parallel computing resources. NWChem has been developed by the Molecular Sciences Software group of the Theory, Modeling & Simulation program of the Environmental Molecular Sciences Laboratory (EMSL) at the Pacific Northwest National Laboratory (PNNL). The early implementation was funded by the EMSL Construction Project.
In mathematics, a graph partition is the reduction of a graph to a smaller graph by partitioning its set of nodes into mutually exclusive groups. Edges of the original graph that cross between the groups will produce edges in the partitioned graph. If the number of resulting edges is small compared to the original graph, then the partitioned graph may be better suited for analysis and problem-solving than the original. Finding a partition that simplifies graph analysis is a hard problem, but one that has applications to scientific computing, VLSI circuit design, and task scheduling in multiprocessor computers, among others. Recently, the graph partition problem has gained importance due to its application for clustering and detection of cliques in social, pathological and biological networks. For a survey on recent trends in computational methods and applications see Buluc et al. (2013). Two common examples of graph partitioning are minimum cut and maximum cut problems.
D. E. Shaw Research (DESRES) is a privately held biochemistry research company based in New York City. Under the scientific direction of David E. Shaw, the group's chief scientist, D. E. Shaw Research develops technologies for molecular dynamics simulations and applies such simulations to basic scientific research in structural biology and biochemistry, and to the process of computer-aided drug design.
Desmond is a software package developed at D. E. Shaw Research to perform high-speed molecular dynamics simulations of biological systems on conventional computer clusters. The code uses novel parallel algorithms and numerical methods to achieve high performance on platforms containing multiple processors, but may also be executed on a single computer.
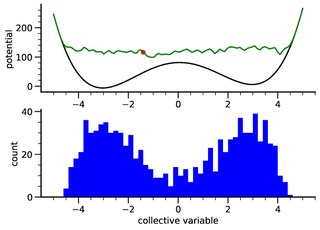
Metadynamics is a computer simulation method in computational physics, chemistry and biology. It is used to estimate the free energy and other state functions of a system, where ergodicity is hindered by the form of the system's energy landscape. It was first suggested by Alessandro Laio and Michele Parrinello in 2002 and is usually applied within molecular dynamics simulations. MTD closely resembles a number of newer methods such as adaptively biased molecular dynamics, adaptive reaction coordinate forces and local elevation umbrella sampling. More recently, both the original and well-tempered metadynamics were derived in the context of importance sampling and shown to be a special case of the adaptive biasing potential setting. MTD is related to the Wang–Landau sampling.
MADNESS is a high-level software environment for the solution of integral and differential equations in many dimensions using adaptive and fast harmonic analysis methods with guaranteed precision based on multiresolution analysis and separated representations .

Christopher Ray Johnson is an American computer scientist. He is a distinguished professor of computer science at the University of Utah, and founding director of the Scientific Computing and Imaging Institute (SCI). His research interests are in the areas of scientific computing and scientific visualization.
Robert J. Harrison is a distinguished expert in high-performance computing. He is a professor in the Applied Mathematics and Statistics department and founding Director of the Institute for Advanced Computational Science at Stony Brook University with a $20M endowment. Through a joint appointment with Brookhaven National Laboratory, Professor Harrison has also been named Director of the Computational Science Center and New York Center for Computational Sciences at Brookhaven. Dr. Harrison comes to Stony Brook from the University of Tennessee and Oak Ridge National Laboratory, where he was Director of the Joint Institute of Computational Science, Professor of Chemistry and Corporate Fellow. He has a prolific career in high-performance computing with over one hundred publications on the subject, as well as extensive service on national advisory committees.
The following is a timeline of scientific computing, also known as computational science.
Model order reduction (MOR) is a technique for reducing the computational complexity of mathematical models in numerical simulations. As such it is closely related to the concept of metamodeling, with applications in all areas of mathematical modelling.
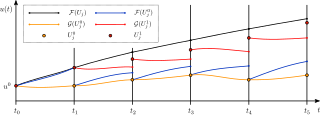
Parareal is a parallel algorithm from numerical analysis and used for the solution of initial value problems. It was introduced in 2001 by Lions, Maday and Turinici. Since then, it has become one of the most widely studied parallel-in-time integration methods.

Haesun Park is a professor and chair of Computational Science and Engineering at the Georgia Institute of Technology. She is an IEEE Fellow, ACM Fellow, and Society for Industrial and Applied Mathematics Fellow. Park's main areas of research are Numerical Algorithms, Data Analysis, Visual Analytics and Parallel Computing. She has co-authored over 100 articles in peer-reviewed journals and conferences.

Ümit V. Çatalyürek is a professor of computer science at the Georgia Institute of Technology, and Adjunct Professor in department of Biomedical Informatics at the Ohio State University. He is known for his work on graph analytics, parallel algorithms for scientific applications, data-intensive computing, and large scale genomic and biomedical applications. He was the director of the High Performance Computing Lab at the Ohio State University. He was named Fellow of the Institute of Electrical and Electronics Engineers (IEEE) in 2016 for contributions to combinatorial scientific computing and parallel computing.

Richard Vuduc is a tenured professor of computer science at the Georgia Institute of Technology. His research lab, The HPC Garage, studies high-performance computing, scientific computing, parallel algorithms, modeling, and engineering. He is a member of the Association for Computing Machinery (ACM). As of 2022, Vuduc serves as Vice President of the SIAM Activity Group on Supercomputing. He has co-authored over 200 articles in peer-reviewed journals and conferences.

Daniel B. Szyld is an Argentinian and American mathematician who is a professor at Temple University in Philadelphia. He has made contributions to numerical and applied linear algebra as well as matrix theory.
Manish Parashar is a Presidential Professor in the School of Computing, Director of the Scientific Computing and Imaging (SCI) Institute and Chair in Computational Science and Engineering at the University of Utah. He also currently serves as Office Director in the US National Science Foundation’s Office of Advanced Cyberinfrastructure. Parashar is the editor-in-chief of IEEE Transactions on Parallel and Distributed Systems, and Founding Chair of the IEEE Technical Community on High Performance Computing. He is an AAAS Fellow, ACM Fellow, and IEEE Fellow.

Computing in Science & Engineering (CiSE) is a bimonthly technical magazine published by the IEEE Computer Society. It was founded in 1999 from the merger of two publications: Computational Science & Engineering (CS&E) and Computers in Physics (CIP), the first published by IEEE and the second by the American Institute of Physics (AIP). The founding editor-in-chief was George Cybenko, known for proving one of the first versions of the universal approximation theorem of neural networks.

Horst D. Simon is a computer scientist known for his contributions to high-performance computing (HPC) and computational science. He is director of ADIA Lab in Abu Dhabi, UAE and editor of TOP500.
References
- ↑ "Edmond Chow's scientific contributions in Parallel and GPU". ResearchGate. Retrieved 2018-01-21.
- 1 2 3 "Edmond Chow". www.cc.gatech.edu. Retrieved 2018-01-21.
- ↑ "Edmond Chow - Google Scholar Citations". scholar.google.com. Retrieved 2018-01-21.
- ↑ "Edmond Chow". dblp.com. Retrieved 2018-01-21.
- ↑ "SIAM Announces Class of 2021 Fellows". March 31, 2021. Retrieved 2021-04-03.
- ↑ Gordon Bell Prize awardees, ACM, accessed 2017-01-21
- ↑ From ‘Superkid’ to supercomputers, East Bay Times, accessed 2017-01-21