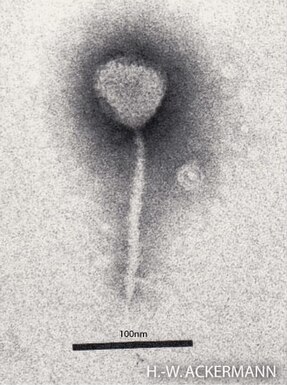
Enterobacteria phage λ is a bacterial virus, or bacteriophage, that infects the bacterial species Escherichia coli. It was discovered by Esther Lederberg in 1950. The wild type of this virus has a temperate life cycle that allows it to either reside within the genome of its host through lysogeny or enter into a lytic phase, during which it kills and lyses the cell to produce offspring. Lambda strains, mutated at specific sites, are unable to lysogenize cells; instead, they grow and enter the lytic cycle after superinfecting an already lysogenized cell.

A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create two identical DNA duplexes from a single original DNA duplex. During this process, DNA polymerase "reads" the existing DNA strands to create two new strands that match the existing ones. These enzymes catalyze the chemical reaction

A nuclease is an enzyme capable of cleaving the phosphodiester bonds between nucleotides of nucleic acids. Nucleases variously affect single and double stranded breaks in their target molecules. In living organisms, they are essential machinery for many aspects of DNA repair. Defects in certain nucleases can cause genetic instability or immunodeficiency. Nucleases are also extensively used in molecular cloning.

The SOS response is a global response to DNA damage in which the cell cycle is arrested and DNA repair and mutagenesis is induced. The system involves the RecA protein. The RecA protein, stimulated by single-stranded DNA, is involved in the inactivation of the repressor (LexA) of SOS response genes thereby inducing the response. It is an error-prone repair system that contributes significantly to DNA changes observed in a wide range of species.

RecBCD is an enzyme of the E. coli bacterium that initiates recombinational repair from potentially lethal double strand breaks in DNA which may result from ionizing radiation, replication errors, endonucleases, oxidative damage, and a host of other factors. The RecBCD enzyme is both a helicase that unwinds, or separates the strands of DNA, and a nuclease that makes single-stranded nicks in DNA.

The nucleoid is an irregularly shaped region within the prokaryotic cell that contains all or most of the genetic material. The chromosome of a prokaryote is circular, and its length is very large compared to the cell dimensions, so it needs to be compacted in order to fit. In contrast to the nucleus of a eukaryotic cell, it is not surrounded by a nuclear membrane. Instead, the nucleoid forms by condensation and functional arrangement with the help of chromosomal architectural proteins and RNA molecules as well as DNA supercoiling. The length of a genome widely varies and a cell may contain multiple copies of it.
RecQ helicase is a family of helicase enzymes initially found in Escherichia coli that has been shown to be important in genome maintenance. They function through catalyzing the reaction ATP + H2O → ADP + P and thus driving the unwinding of paired DNA and translocating in the 3' to 5' direction. These enzymes can also drive the reaction NTP + H2O → NDP + P to drive the unwinding of either DNA or RNA.

Exonucleases are enzymes that work by cleaving nucleotides one at a time from the end (exo) of a polynucleotide chain. A hydrolyzing reaction that breaks phosphodiester bonds at either the 3′ or the 5′ end occurs. Its close relative is the endonuclease, which cleaves phosphodiester bonds in the middle (endo) of a polynucleotide chain. Eukaryotes and prokaryotes have three types of exonucleases involved in the normal turnover of mRNA: 5′ to 3′ exonuclease (Xrn1), which is a dependent decapping protein; 3′ to 5′ exonuclease, an independent protein; and poly(A)-specific 3′ to 5′ exonuclease.

RecA is a 38 kilodalton protein essential for the repair and maintenance of DNA. A RecA structural and functional homolog has been found in every species in which one has been seriously sought and serves as an archetype for this class of homologous DNA repair proteins. The homologous protein is called RAD51 in eukaryotes and RadA in archaea.

Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids. It is widely used by cells to accurately repair harmful breaks that occur on both strands of DNA, known as double-strand breaks (DSB), in a process called homologous recombinational repair (HRR). Homologous recombination also produces new combinations of DNA sequences during meiosis, the process by which eukaryotes make gamete cells, like sperm and egg cells in animals. These new combinations of DNA represent genetic variation in offspring, which in turn enables populations to adapt during the course of evolution. Homologous recombination is also used in horizontal gene transfer to exchange genetic material between different strains and species of bacteria and viruses.
Recombineering is a genetic and molecular biology technique based on homologous recombination systems, as opposed to the older/more common method of using restriction enzymes and ligases to combine DNA sequences in a specified order. Recombineering is widely used for bacterial genetics, in the generation of target vectors for making a conditional mouse knockout, and for modifying DNA of any source often contained on a bacterial artificial chromosome (BAC), among other applications.
Recombinases are genetic recombination enzymes.
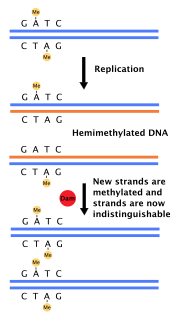
DNA adenine methylase, (Dam methylase) is an enzyme that adds a methyl group to the adenine of the sequence 5'-GATC-3' in newly synthesized DNA. Immediately after DNA synthesis, the daughter strand remains unmethylated for a short time. It is an orphan methyltransferase that is not part of a restriction-modification system and regulates gene expression. This enzyme catalyses the following chemical reaction
Prokaryotic DNA Replication is the process by which a prokaryote duplicates its DNA into another copy that is passed on to daughter cells. Although it is often studied in the model organism E. coli, other bacteria show many similarities. Replication is bi-directional and originates at a single origin of replication (OriC). It consists of three steps: Initiation, elongation, and termination.
Deoxyribonuclease IV (phage-T4-induced) is a kind of Endonuclease that catalyzes the degradation nucleotides in DsDNA by attacking the 5'-terminal end.
The RecF pathway, also called the RecFOR pathway, is a pathway of homologous recombination that repairs DNA in bacteria. It repairs breaks that occur on only one of DNA's two strands, known as single-strand gaps. The RecF pathway can also repair double-strand breaks in DNA when the RecBCD pathway, another pathway of homologous recombination in bacteria, is inactivated by mutations. Like the RecBCD pathway, the RecF pathway requires RecA for strand invasion. The two pathways are also similar in their phases of branch migration, in which the Holliday junction slides in one direction, and resolution, in which the Holliday junctions are cleaved apart by enzymes.
In molecular biology, bacterial DNA binding proteins are a family of small, usually basic proteins of about 90 residues that bind DNA and are known as histone-like proteins. Since bacterial binding proteins have a diversity of functions, it has been difficult to develop a common function for all of them. They are commonly referred to as histone-like and have many similar traits with the eukaryotic histone proteins. Eukaryotic histones package DNA to help it to fit in the nucleus, and they are known to be the most conserved proteins in nature. Examples include the HU protein in Escherichia coli, a dimer of closely related alpha and beta chains and in other bacteria can be a dimer of identical chains. HU-type proteins have been found in a variety of bacteria and archaea, and are also encoded in the chloroplast genome of some algae. The integration host factor (IHF), a dimer of closely related chains which is suggested to function in genetic recombination as well as in translational and transcriptional control is found in Enterobacteria and viral proteins including the African swine fever virus protein A104R.
DNA Polymerase V is a polymerase enzyme involved in DNA repair mechanisms in bacteria, such as Escherichia coli. It is composed of a UmuD' homodimer and a UmuC monomer, forming the UmuD'2C protein complex. It is part of the Y-family of DNA Polymerases, which are capable of performing DNA translesion synthesis (TLS). Translesion polymerases bypass DNA damage lesions during DNA replication - if a lesion is not repaired or bypassed the replication fork can stall and lead to cell death. However, Y polymerases have low sequence fidelity during replication. When the UmuC and UmuD' proteins were initially discovered in E. coli, they were thought to be agents that inhibit faithful DNA replication and caused DNA synthesis to have high mutation rates after exposure to UV-light. The polymerase function of Pol V was not discovered until the late 1990s when UmuC was successfully extracted, consequent experiments unequivocally proved UmuD'2C is a polymerase. This finding lead to the detection of many Pol V orthologs and the discovery of the Y-family of polymerases.
SCAR-less genome editing Scarless Cas9 Assisted Recombineering (no-SCAR) is an editing method that is able to manipulate the Escherichia coli genome. The system relies on recombineering whereby DNA sequences are combined and manipulated through homologous recombination. No-SCAR is able to manipulate the E. coli genome without the use of the chromosomal markers detailed in previous recombineering methods. Instead, in this method, the λ-Red recombination system facilitates donor DNA integration while Cas9 cleaves double-stranded DNA to counter-select against wild-type cells. Although λ-Red and Cas9 genome editing are widely used technologies, the no-SCAR method is novel in combining the two functions; this technique is able to establish point mutations, gene deletions, and short sequence insertions in several genomic loci with increased efficiency and time sensitivity.
Susan Thomas Lovett is an American molecular biologist who is the Abraham S. and Gertrude Burg Professor of Microbiology at Brandeis University. She is interested in the mechanisms that allow the genetic material in cells to remain stable over time.












