P elements are transposable elements that were discovered in Drosophila as the causative agents of genetic traits called hybrid dysgenesis. The transposon is responsible for the P trait of the P element and it is found only in wild flies. They are also found in many other eukaryotes.

In molecular biology, SNORA66 is a non-coding RNA (ncRNA) molecule which functions in the biogenesis (modification) of other small nuclear RNAs (snRNAs). This type of modifying RNA is located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a "guide RNA". U66 was originally cloned from HeLa cells and belongs to the H/ACA box class of snoRNAs as it has the predicted hairpin-hinge-hairpin-tail structure, has the conserved H/ACA-box motifs and is found associated with GAR1 protein. U66 is predicted to guide the pseudouridylation of U119 of 18S ribosomal RNA (rRNA). Pseudouridylation is the to the different isomeric form pseudouridine.

In molecular biology, Small nucleolar RNA SNORA68 is a non-coding RNA (ncRNA) molecule which functions in the biogenesis (modification) of other small nuclear RNAs (snRNAs). This type of modifying RNA is located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a "guide RNA".

In molecular biology, Small nucleolar RNA SNORA69 is a non-coding RNA (ncRNA) molecule which functions in the biogenesis (modification) of other small nuclear RNAs (snRNAs). This type of modifying RNA is located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a "guide RNA".

In molecular biology, small nucleolar RNA SNORA72 is a non-coding RNA (ncRNA) molecule which functions in the biogenesis (modification) of other small nuclear RNAs (snRNAs). This type of modifying RNA is located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a "guide RNA".

In molecular biology, Small nucleolar RNA SNORA70 is a non-coding RNA (ncRNA) molecule which functions in the biogenesis (modification) of other small nuclear RNAs (snRNAs). This type of modifying RNA is located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a "guide RNA".
Iterons are directly repeated DNA sequences which play an important role in regulation of plasmid copy number in bacterial cells. It is one among the three negative regulatory elements found in plasmids which control its copy number. The others include antisense RNAs and ctRNAs. Iterons complex with cognate replication (Rep) initiator proteins to achieve the required regulatory effect.
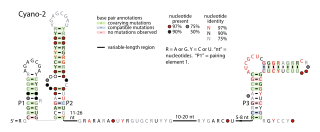
The Cyano-2 RNA motif is a conserved RNA structure identified by bioinformatics. Cyano-2 RNAs are found in Cyanobacterial species classified within the genus Synechococcus. Many terminal loops in the two conserved stem-loops contain the nucleotide sequence GCGA, and these sequences might in some cases form stable GNRA tetraloops. Since the two stem-loops are somewhat distant from one another it is possible that they represent two independent non-coding RNAs that are often or always co-transcribed. The region one thousand base pairs upstream of predicted Cyano-2 RNAs is usually devoid of annotated features such as RNA or protein-coding genes. This absence of annotated genes within one thousand base pairs is relatively unusual within bacteria.
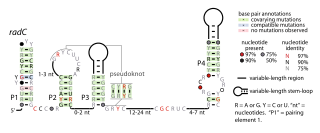
The radC RNA motif is a conserved RNA structure identified by bioinformatics. The radC RNA motif is found in certain bacteria where it is consistent located in the presumed 5' untranslated regions of genes whose encoded proteins bind DNA are interact with other proteins that bind DNA. These proteins include integrases, methyltransferases that might methylate DNA, proteins that inhibit restriction enzymes and radC genes. Although radC genes were thought to encode DNA repair proteins, this conclusion was based on mutation data that was later shown to affect a different gene. However, it is still possible that radC genes play some DNA-related role. No radC RNAs have been detected in any purified phage whose sequence was available as of 2010, although integrases are often used by phages.

The Ssbp, Topoisomerase, Antirestriction, XerDC Integrase RNA motif is a conserved RNA-like structure identified using bioinformatics. STAXI RNAs are located near to genes encoding proteins that interact with DNA or are associated with such proteins. This observation raised the possibility that instances of the STAXI motif function as single-stranded DNA molecules, perhaps during DNA replication or DNA repair. On the other hand, STAXI motifs often contain terminal loops conforming to the stable UNCG tetraloop, but the DNA version of this tetraloop (TNCG) is not especially stable. The STAXI motif consists of a simple pseudoknot structure that is repeated two or more times.

The traJ-II RNA motif is a conserved RNA structure discovered in bacteria by using bioinformatics. traJ-II RNAs appear to be in the 5' untranslated regions of protein-coding genes called traJ, which functions in the process of bacterial conjugation. A previously identified motif known as TraJ 5' UTR is also found upstream of traJ genes functions as the target of FinP antisense RNAs, so it is possible that traJ-II RNAs play a similar role as targets of an antisense RNA. However, some sequence features within the traJ-II RNA motif suggest that the biological RNA might be transcribed from the reverse-complement strand. Thus is it unclear whether traJ-II function as cis-regulatory elements. traJ-II RNAs are found in a variety of Pseudomonadota.
αr35 is a family of bacterial small non-coding RNAs with representatives in a reduced group of Alphaproteobacteria from the order Hyphomicrobiales. The first member of this family (Smr35B) was found in a Sinorhizobium meliloti 1021 locus located in the symbiotic plasmid B (pSymB). Further homology and structure conservation analysis have identified full-length SmrB35 homologs in other legume symbionts, as well as in the human and plant pathogens Brucella anthropi and Agrobacterium tumefaciens, respectively. αr35 RNA species are 139-142 nt long and share a common secondary structure consisting of two stem loops and a well conserved rho independent terminator. Most of the αr35 transcripts can be catalogued as trans-acting sRNAs expressed from well-defined promoter regions of independent transcription units within intergenic regions of the Alphaproteobacterial genomes.

The ARRPOF RNA motif is a conserved RNA structure that was discovered by bioinformatics.

The aspS RNA motif is a conserved RNA structure that was discovered by bioinformatics. aspS motifs are found in a specific lineage of Actinomycetota.
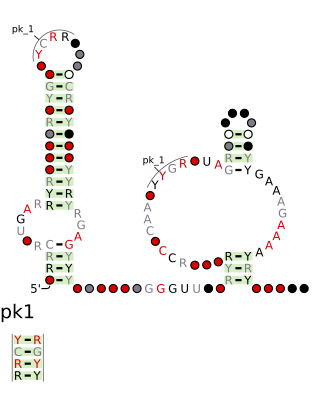
The drum RNA motif is a conserved RNA structure that was discovered by bioinformatics. Drum motifs are found in Bacillota, Bacteroidota, Pseudomonadota, and Spirochaetota, and exhibit multiple highly conserved nucleotide positions, despite their widespread distribution.

The DUF2800 RNA motif is a conserved RNA structure that was discovered by bioinformatics. DUF2800 motif RNAs are found in Bacillota. DUF2800 RNAs are also predicted in the phyla Actinomycetota and Synergistota, although these RNAs are likely the result of recent horizontal gene transfer or conceivably sequence contamination.

The DUF3577 RNA motif is a conserved RNA structure that was discovered by bioinformatics. DUF3577 motifs are found in the organism Cardiobacterium valvarum and metagenomic sequences from unknown organisms.

The Extended-Gap, in Firmicutes and One Actiobacterium RNA motif is a conserved RNA structure that was discovered by bioinformatics. EGFOA motifs are found in Bacillota and one example was detected in Actinomycetota. The EGFOA-assoc-1 and EGFOA-assoc-2 RNA motifs are conserved RNA structures that are often located nearby to EGFOA RNAs, and presumably functions in some related mechanism.

The Enterococcus-1 RNA motif is a conserved RNA structure that was discovered by bioinformatics. Enterococcus-1 motif RNAs are found in bacteria of the genus Enterococcus.
The freshwater-2 RNA motif is a conserved RNA structure that was discovered by bioinformatics. Freshwater-2 motif RNAs are found in metagenomic sequences that are isolated from aquatic and especially freshwater environments. As of 2018, no freshwater-2 RNA has been identified in a classified organism.
















