
In genetics and bioinformatics, a single-nucleotide polymorphism is a germline substitution of a single nucleotide at a specific position in the genome that is present in a sufficiently large fraction of the population.
Heterosis, hybrid vigor, or outbreeding enhancement is the improved or increased function of any biological quality in a hybrid offspring. An offspring is heterotic if its traits are enhanced as a result of mixing the genetic contributions of its parents. The heterotic offspring often has traits that are more than the simple addition of the parents' traits, and can be explained by Mendelian or non-Mendelian inheritance. Typical heterotic/hybrid traits of interest in agriculture are higher yield, quicker maturity, stability, drought tolerance etc.

The laboratory mouse or lab mouse is a small mammal of the order Rodentia which is bred and used for scientific research or feeders for certain pets. Laboratory mice are usually of the species Mus musculus. They are the most commonly used mammalian research model and are used for research in genetics, physiology, psychology, medicine and other scientific disciplines. Mice belong to the Euarchontoglires clade, which includes humans. This close relationship, the associated high homology with humans, their ease of maintenance and handling, and their high reproduction rate, make mice particularly suitable models for human-oriented research. The laboratory mouse genome has been sequenced and many mouse genes have human homologues. Lab mice sold at pet stores for snake food can also be kept as pets.
The Human Genome Diversity Project (HGDP) was started by Stanford University's Morrison Institute in 1990s along with collaboration of scientists around the world. It is the result of many years of work by Luigi Cavalli-Sforza, one of the most cited scientists in the world, who has published extensively in the use of genetics to understand human migration and evolution. The HGDP data sets have often been cited in papers on such topics as population genetics, anthropology, and heritable disease research.
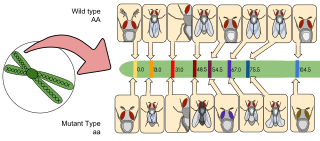
Gene mapping or genome mapping describes the methods used to identify the location of a gene on a chromosome and the distances between genes. Gene mapping can also describe the distances between different sites within a gene.

Introgression, also known as introgressive hybridization, in genetics is the transfer of genetic material from one species into the gene pool of another by the repeated backcrossing of an interspecific hybrid with one of its parent species. Introgression is a long-term process, even when artificial; it may take many hybrid generations before significant backcrossing occurs. This process is distinct from most forms of gene flow in that it occurs between two populations of different species, rather than two populations of the same species.

Human genetic variation is the genetic differences in and among populations. There may be multiple variants of any given gene in the human population (alleles), a situation called polymorphism.

In genomics, a genome-wide association study, is an observational study of a genome-wide set of genetic variants in different individuals to see if any variant is associated with a trait. GWA studies typically focus on associations between single-nucleotide polymorphisms (SNPs) and traits like major human diseases, but can equally be applied to any other genetic variants and any other organisms.

The 1000 Genomes Project, launched in January 2008, was an international research effort to establish by far the most detailed catalogue of human genetic variation. Scientists planned to sequence the genomes of at least one thousand anonymous participants from a number of different ethnic groups within the following three years, using newly developed technologies which were faster and less expensive. In 2010, the project finished its pilot phase, which was described in detail in a publication in the journal Nature. In 2012, the sequencing of 1092 genomes was announced in a Nature publication. In 2015, two papers in Nature reported results and the completion of the project and opportunities for future research.
Population genomics is the large-scale comparison of DNA sequences of populations. Population genomics is a neologism that is associated with population genetics. Population genomics studies genome-wide effects to improve our understanding of microevolution so that we may learn the phylogenetic history and demography of a population.
Genetic admixture occurs when previously diverged or isolated genetic lineages mix. Admixture results in the introduction of new genetic lineages into a population.

Behavioural genetics, also referred to as behaviour genetics, is a field of scientific research that uses genetic methods to investigate the nature and origins of individual differences in behaviour. While the name "behavioural genetics" connotes a focus on genetic influences, the field broadly investigates the extent to which genetic and environmental factors influence individual differences, and the development of research designs that can remove the confounding of genes and environment. Behavioural genetics was founded as a scientific discipline by Francis Galton in the late 19th century, only to be discredited through association with eugenics movements before and during World War II. In the latter half of the 20th century, the field saw renewed prominence with research on inheritance of behaviour and mental illness in humans, as well as research on genetically informative model organisms through selective breeding and crosses. In the late 20th and early 21st centuries, technological advances in molecular genetics made it possible to measure and modify the genome directly. This led to major advances in model organism research and in human studies, leading to new scientific discoveries.
Expression quantitative trait loci (eQTLs) are genomic loci that explain variation in expression levels of mRNAs.
In genetics, association mapping, also known as "linkage disequilibrium mapping", is a method of mapping quantitative trait loci (QTLs) that takes advantage of historic linkage disequilibrium to link phenotypes to genotypes, uncovering genetic associations.
Nested association mapping (NAM) is a technique designed by the labs of Edward Buckler, James Holland, and Michael McMullen for identifying and dissecting the genetic architecture of complex traits in corn. It is important to note that nested association mapping is a specific technique that cannot be performed outside of a specifically designed population such as the Maize NAM population, the details of which are described below.
GeneNetwork is a combined database and open-source bioinformatics data analysis software resource for systems genetics. This resource is used to study gene regulatory networks that link DNA sequence differences to corresponding differences in gene and protein expression and to variation in traits such as health and disease risk. Data sets in GeneNetwork are typically made up of large collections of genotypes and phenotypes from groups of individuals, including humans, strains of mice and rats, and organisms as diverse as Drosophila melanogaster, Arabidopsis thaliana, and barley. The inclusion of genotypes makes it practical to carry out web-based gene mapping to discover those regions of genomes that contribute to differences among individuals in mRNA, protein, and metabolite levels, as well as differences in cell function, anatomy, physiology, and behavior.
Domesticated species and the human populations that domesticate them are typified by a mutualistic relationship of interdependence, in which humans have over thousands of years modified the genomics of domesticated species. Genomics is the study of the structure, content, and evolution of genomes, or the entire genetic information of organisms. Domestication is the process by which humans alter the morphology and genes of targeted organisms by selecting for desirable traits. These genomic changes produce the domestication syndromes.

In genetics, a polygenic score (PGS), also called a polygenic index (PGI), polygenic risk score (PRS), genetic risk score, or genome-wide score, is a number that summarizes the estimated effect of many genetic variants on an individual's phenotype, typically calculated as a weighted sum of trait-associated alleles. It reflects an individual's estimated genetic predisposition for a given trait and can be used as a predictor for that trait. In other words, it gives an estimate of how likely an individual is to have a given trait only based on genetics, without taking environmental factors into account. Polygenic scores are widely used in animal breeding and plant breeding due to their efficacy in improving livestock breeding and crops. In humans, polygenic scores are typically generated from genome-wide association study (GWAS) data.

Complex traits, also known as quantitative traits, are traits that do not behave according to simple Mendelian inheritance laws. More specifically, their inheritance cannot be explained by the genetic segregation of a single gene. Such traits show a continuous range of variation and are influenced by both environmental and genetic factors. Compared to strictly Mendelian traits, complex traits are far more common, and because they can be hugely polygenic, they are studied using statistical techniques such as quantitative genetics and quantitative trait loci (QTL) mapping rather than classical genetics methods. Examples of complex traits include height, circadian rhythms, enzyme kinetics, and many diseases including diabetes and Parkinson's disease. One major goal of genetic research today is to better understand the molecular mechanisms through which genetic variants act to influence complex traits.

Edward S. Buckler is a plant geneticist with the USDA Agricultural Research Service and holds an adjunct appointment at Cornell University. His work focuses on both quantitative and statistical genetics in maize as well as other crops such as cassava. He originated the concept of Nested association mapping and created the first population designed for this type of quantitative genetic analysis. Buckler was elected an American Association for the Advancement of Science Fellow in 2012. In 2014, he was elected to the National Academy of Sciences. In 2017, he received the NAS prize in Food and Agricultural Science for his work using natural genetic diversity to develop varieties of maize with fifteen times more vitamin A than existing varieties.










