General research
Research by Arron et al. shows that some of the phenotypes associated with Down syndrome can be related to the disregulation of transcription factors (596), and in particular, NFAT. NFAT is controlled in part by two proteins, DSCR1 and DYRK1A; these genes are located on chromosome-21 (Epstein 582). In people with Down syndrome, these proteins have 1.5 times greater concentration than normal (Arron et al. 597). The elevated levels of DSCR1 and DYRK1A keep NFAT primarily located in the cytoplasm rather than in the nucleus, preventing NFATc from activating the transcription of target genes and thus the production of certain proteins (Epstein 583).
This disregulation was discovered by testing in transgenic mice that had segments of their chromosomes duplicated to simulate a human chromosome-21 trisomy (Arron et al. 597). A test involving grip strength showed that the genetically modified mice had a significantly weaker grip, much like the characteristically poor muscle tone of an individual with Down syndrome (Arron et al. 596). The mice squeezed a probe with a paw and displayed a 0.2 newton weaker grip (Arron et al. 596). Down syndrome is also characterized by increased socialization. When modified and unmodified mice were observed for social interaction, the modified mice showed as much as 25% more interactions as compared to the unmodified mice (Arron et al. 596).
The genes that may be responsible for the phenotypes associated may be located proximal to 21q22.3. Testing by Olson and others in transgenic mice show the duplicated genes presumed to cause the phenotypes are not enough to cause the exact features. While the mice had sections of multiple genes duplicated to approximate a human chromosome-21 triplication, they only showed slight craniofacial abnormalities (688–90). The transgenic mice were compared to mice that had no gene duplication by measuring distances on various points on their skeletal structure and comparing them to the normal mice (Olson et al. 687). The exact characteristics of Down syndrome were not observed, so more genes involved for Down syndrome phenotypes have to be located elsewhere.
Reeves et al., using 250 clones of chromosome-21 and specific gene markers, were able to map the gene in mutated bacteria. The testing had 99.7% coverage of the gene with 99.9995% accuracy due to multiple redundancies in the mapping techniques. In the study 225 genes were identified (311–13).
The search for major genes that may be involved in Down syndrome symptoms is normally in the region 21q21–21q22.3. However, studies by Reeves et al. show that 41% of the genes on chromosome-21 have no functional purpose, and only 54% of functional genes have a known protein sequence. Functionality of genes was determined by a computer using exon prediction analysis (312). Exon sequence was obtained by the same procedures of the chromosome-21 mapping.
Research has led to an understanding that two genes located on chromosome-21, that code for proteins that control gene regulators, DSCR1 and DYRK1A can be responsible for some of the phenotypes associated with Down syndrome. DSCR1 and DYRK1A cannot be blamed outright for the symptoms; there are a lot of genes that have no known purpose. Much more research would be needed to produce any appropriate or ethically acceptable treatment options.
Recent use of transgenic mice to study specific genes in the Down syndrome critical region has yielded some results. APP [10] is an Amyloid beta A4 precursor protein. It is suspected to have a major role in cognitive difficulties. [11] Another gene, ETS2 [12] is Avian Erythroblastosis Virus E26 Oncogene Homolog 2. Researchers have "demonstrated that over-expression of ETS2 results in apoptosis. Transgenic mice over-expressing ETS2 developed a smaller thymus and lymphocyte abnormalities, similar to features observed in Down syndrome." [12]
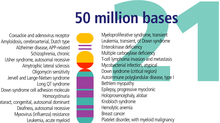
Chromosome 21 is one of the 23 pairs of chromosomes in humans. Chromosome 21 is both the smallest human autosome and chromosome, with 45 million base pairs representing about 1.5 percent of the total DNA in cells. Most people have two copies of chromosome 21, while those with three copies of chromosome 21 have Down syndrome, also called "trisomy 21".

Agouti-signaling protein is a protein that in humans is encoded by the ASIP gene. It is responsible for the distribution of melanin pigment in mammals. Agouti interacts with the melanocortin 1 receptor to determine whether the melanocyte produces phaeomelanin, or eumelanin. This interaction is responsible for making distinct light and dark bands in the hairs of animals such as the agouti, which the gene is named after. In other species such as horses, agouti signalling is responsible for determining which parts of the body will be red or black. Mice with wildtype agouti will be grey, with each hair being partly yellow and partly black. Loss of function mutations in mice and other species cause black fur coloration, while mutations causing expression throughout the whole body in mice cause yellow fur and obesity.

The autoimmune regulator (AIRE) is a protein that in humans is encoded by the AIRE gene. It is a 13kb gene on chromosome 21q22.3 that has 545 amino acids. AIRE is a transcription factor expressed in the medulla of the thymus. It is part of the mechanism which eliminates self-reactive T cells that would cause autoimmune disease. It exposes T cells to normal, healthy proteins from all parts of the body, and T cells that react to those proteins are destroyed.

Trifunctional enzyme subunit beta, mitochondrial (TP-beta) also known as 3-ketoacyl-CoA thiolase, acetyl-CoA acyltransferase, or beta-ketothiolase is an enzyme that in humans is encoded by the HADHB gene.

Superoxide dismutase [Cu-Zn] also known as superoxide dismutase 1 or hSod1 is an enzyme that in humans is encoded by the SOD1 gene, located on chromosome 21. SOD1 is one of three human superoxide dismutases. It is implicated in apoptosis, familial amyotrophic lateral sclerosis and Parkinson's disease.

Protein C-ETS2 is a protein that in humans is encoded by the ETS2 gene. The protein encoded by this gene belongs to the ETS family of transcription factors.

Dual specificity tyrosine-phosphorylation-regulated kinase 1A is an enzyme that in humans is encoded by the DYRK1A gene. Alternative splicing of this gene generates several transcript variants differing from each other either in the 5' UTR or in the 3' coding region. These variants encode at least five different isoforms.

Nuclear factor of activated T-cells 5, also known as NFAT5 and sometimes TonEBP, is a human gene that encodes a transcription factor that regulates the expression of genes involved in the osmotic stress.

Down syndrome critical region gene 1, also known as DSCR1, is a protein that in humans is encoded by the RCAN1 gene.

Carbonyl reductase 1, also known as CBR1, is an enzyme which in humans is encoded by the CBR1 gene. The protein encoded by this gene belongs to the short-chain dehydrogenases/reductases (SDR) family, which function as NADPH-dependent oxidoreductases having wide specificity for carbonyl compounds, such as quinones, prostaglandins, and various xenobiotics. Alternatively spliced transcript variants have been found for this gene.

Paired box protein Pax-1 is a protein that in humans is encoded by the PAX1 gene.

PR domain containing 16, also known as PRDM16, is a protein which in humans is encoded by the PRDM16 gene.

The LBH gene is a highly conserved human gene that produces the LBH protein, a transcription co-factor in the Wnt/β-catenin pathway. Upon transcriptional activation of β-catenin, LBH goes on to act as a regulator of cell proliferation and differentiation through multiple transcriptional targets. The gene is located on the p arm of chromosome 2 and is roughly 28 kb long. Current ongoing studies are examining its role in developmental and oncological settings.

DOP1B is a human gene located just above the Down Syndrome chromosomal region (DSCR) located at 21p22.2 sub-band. Although the exact function of this gene is not yet fully understood, it has been proven to play a role in multiple biological processes, and its over-expression (triplication) has been linked to multiple facets of the Down Syndrome phenotype, most notably mental retardation.

tRNA -methyltransferase subunit WDR4 is an enzyme that in humans is encoded by the WDR4 gene.

Tetratricopeptide repeat protein 3 is a protein that in humans is encoded by the TTC3 gene.

Subunit P of phosphatidylinositol N-acetylglucosaminyltransferase is an enzyme that in humans is encoded by the PIGP gene.

RCAN3 is a gene that in humans encodes the Calcipressin-3 protein.
The DNA damage theory of aging proposes that aging is a consequence of unrepaired accumulation of naturally occurring DNA damage. Damage in this context is a DNA alteration that has an abnormal structure. Although both mitochondrial and nuclear DNA damage can contribute to aging, nuclear DNA is the main subject of this analysis. Nuclear DNA damage can contribute to aging either indirectly or directly.
Mouse models have frequently been used to study Down syndrome due to the close similarity in the genomes of mice and humans, and the prevalence of mice usage in laboratory research.
This page is based on this
Wikipedia article Text is available under the
CC BY-SA 4.0 license; additional terms may apply.
Images, videos and audio are available under their respective licenses.