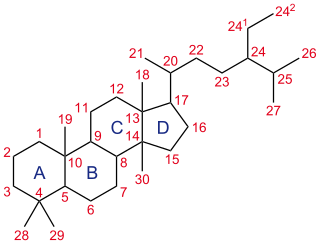
A steroid is a biologically active organic compound with four rings arranged in a specific molecular configuration. Steroids have two principal biological functions: as important components of cell membranes that alter membrane fluidity; and as signaling molecules. Hundreds of steroids are found in plants, animals and fungi. All steroids are manufactured in cells from the sterols lanosterol (opisthokonts) or cycloartenol (plants). Lanosterol and cycloartenol are derived from the cyclization of the triterpene squalene.

Lanosterol synthase is an oxidosqualene cyclase (OSC) enzyme that converts (S)-2,3-oxidosqualene to a protosterol cation and finally to lanosterol. Lanosterol is a key four-ringed intermediate in cholesterol biosynthesis. In humans, lanosterol synthase is encoded by the LSS gene.
In enzymology, a Delta24(241)-sterol reductase (EC 1.3.1.71) is an enzyme that catalyzes the chemical reaction
In enzymology, a Δ7-sterol 5(6)-desaturase is an enzyme that catalyzes the chemical reaction
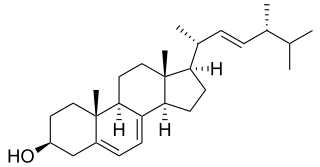
In enzymology, a sterol 14-demethylase (EC 1.14.13.70) is an enzyme of the Cytochrome P450 (CYP) superfamily. It is any member of the CYP51 family. It catalyzes a chemical reaction such as:
Sterol O-acyltransferase is an intracellular protein located in the endoplasmic reticulum that forms cholesteryl esters from cholesterol.
The oxysterol-binding protein (OSBP)-related proteins (ORPs) are a family of lipid transfer proteins (LTPs). Concretely, they constitute a family of sterol and phosphoinositide binding and transfer proteins in eukaryotes that are conserved from yeast to humans. They are lipid-binding proteins implicated in many cellular processes related with oxysterol, including signaling, vesicular trafficking, lipid metabolism, and nonvesicular sterol transport.

Lathosterol oxidase is a Δ7-sterol 5(6)-desaturase enzyme that in humans is encoded by the SC5D gene.
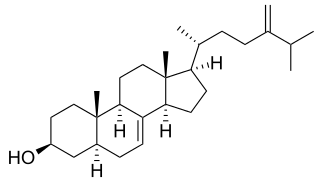
Episterol is a sterol involved in the biosynthesis of steroids. Episterol is converted from 24-methylenelophenol. Episterol is converted to 5-dehydroepisterol by ERG3, the C-5 sterol desaturase in the yeast. Episterol is also known to be a precursor to ergosterol.
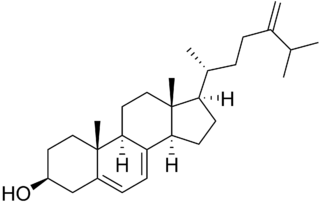
5-Dehydroepisterol is a sterol and an intermediate in steroid biosynthesis, particularly synthesis of brassinosteroids. It is formed from episterol through action of ERG3, the C-5 sterol desaturase in the yeast and is then converted into 24-methylenecholesterol by 7-dehydrocholesterol reductase.
Membrane contact sites (MCS) are close appositions between two organelles. Ultrastructural studies typically reveal an intermembrane distance in the order of the size of a single protein, as small as 10 nm or wider, with no clear upper limit. These zones of apposition are highly conserved in evolution. These sites are thought to be important to facilitate signalling, and they promote the passage of small molecules, including ions, lipids and reactive oxygen species. MCS are important in the function of the endoplasmic reticulum (ER), since this is the major site of lipid synthesis within cells. The ER makes close contact with many organelles, including mitochondria, Golgi, endosomes, lysosomes, peroxisomes, chloroplasts and the plasma membrane. Both mitochondria and sorting endosomes undergo major rearrangements leading to fission where they contact the ER. Sites of close apposition can also form between most of these organelles most pairwise combinations. First mentions of these contact sites can be found in papers published in the late 1950s mainly visualized using electron microscopy (EM) techniques. Copeland and Dalton described them as “highly specialized tubular form of endoplasmic reticulum in association with the mitochondria and apparently in turn, with the vascular border of the cell”.

Sec14 is a cytosolic protein found in yeast which plays a role in the regulation of several cellular functions, specifically those related to intracellular transport. Encoded by the Sec14 gene, Sec14p may transport phosphatidylinositol and phosphatidylcholine produced in the endoplasmic reticulum and the Golgi body to other cellular membranes. Additionally, Sec14p potentially plays a role in the localization of lipid raft proteins. Sec14p is an essential gene in yeast, and is homologous in function to phosphatidylinositol transfer protein in mammals. A conditional mutant with non-functional Sec14p presents with Berkeley bodies and deficiencies in protein secretion.
Stearoyl-CoA is a coenzyme involved in the metabolism of fatty acids. Stearoyl-CoA is an 18-carbon long fatty acyl-CoA chain that participates in an unsaturation reaction. The reaction is catalyzed by the enzyme stearoyl-CoA desaturase, which is located in the endoplasmic reticulum. It forms a cis-double bond between the ninth and tenth carbons within the chain to form the product oleoyl-CoA.

C-5 sterol desaturase is an enzyme that is highly conserved among eukaryotes and catalyzes the dehydrogenation of a C-5(6) bond in a sterol intermediate compound as a step in the biosynthesis of major sterols. The precise structure of the enzyme’s substrate varies by species. For example, the human C-5 sterol desaturase oxidizes lathosterol, while its ortholog ERG3 in the yeast Saccharomyces cerevisiae oxidizes episterol.

Methylsterol monooxygenase 1 is a protein that in humans is encoded by the MSMO1 gene.
ERG11 or Sterol 14-demethylase is a fungal cytochrome P450 enzyme originally from Saccharomyces cerevisiae, belongs to family CYP51, with the CYP Symbol CYP51F1. ERG11 catalyzes the C14-demethylation of lanosterol to 4,4'-dimethyl cholesta-8,14,24-triene-3-beta-ol which is the first step of biosynthesis of the zymosterol, zymosterol will be further converted into Ergosterol.
ERG5 or Sterol 22-desaturase is a cytochrome P450 enzyme in the ergosterol biosynthesis pathway of fungi Saccharomyces cerevisiae, with the CYP Symbol CYP61A1. CYP61A1 is one of only three P450 enzyme found in baker's yeast, the other two are CYP51F1 and CYP56A1. The ortholog in Schizosaccharomyces pombe, was named CYP61A3 for historical reasons, and is only one of two P450 enzyme found with CYP51F1. ERG5 catalyzes the C22-C23 double bond formation on the sterol side chain of ergostatrienol to convert it into ergostatetraenol, then the C24 double bond of ergostatetrenol will be hydrogenation reduced into ergosterol by ERG4.
Cytochrome P450-DIT2 or CYP56A1 is one of the only three P450 enzyme found in fungi baker's yeast, the other two are CYP51F1(ERG11) and CYP61A1(ERG5) in the ergosterol biosynthesis pathway. CYP56A1 thought to catalyze the oxidation of tyrosine residues in the formation of L,L-dityrosine a precursor of the spore wall.
ERG4 or Delta(24 )-sterol reductase or Delta(24 )-sterol reductase is an enzyme that catalyzes the last step of ergosterol biosynthesis pathway in fungi Saccharomyces cerevisiae, which 5,7,22,24(28)-ergostatetraenol converted into ergosterol.

24-Methylenelophenol, or Gramisterol, also called 4α-Methyl-5α-ergosta-7,24(28)-dien-3β-ol is a Metabolic intermediate of sterol biosynthesis of plants and fungis, can be converted from 4α-Methylfecosterol by enzyme HYD1 and converted to (Z)-24-ethylidenelophenol by 24-methylenesterol C-methyltransferase.












