Related Research Articles

Endocytosis is a cellular process in which substances are brought into the cell. The material to be internalized is surrounded by an area of cell membrane, which then buds off inside the cell to form a vesicle containing the ingested materials. Endocytosis includes pinocytosis and phagocytosis. It is a form of active transport.

Endosomes are a collection of intracellular sorting organelles in eukaryotic cells. They are parts of endocytic membrane transport pathway originating from the trans Golgi network. Molecules or ligands internalized from the plasma membrane can follow this pathway all the way to lysosomes for degradation or can be recycled back to the cell membrane in the endocytic cycle. Molecules are also transported to endosomes from the trans Golgi network and either continue to lysosomes or recycle back to the Golgi apparatus.
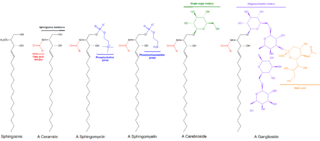
Sphingolipids are a class of lipids containing a backbone of sphingoid bases, which are a set of aliphatic amino alcohols that includes sphingosine. They were discovered in brain extracts in the 1870s and were named after the mythological sphinx because of their enigmatic nature. These compounds play important roles in signal transduction and cell recognition. Sphingolipidoses, or disorders of sphingolipid metabolism, have particular impact on neural tissue. A sphingolipid with a terminal hydroxyl group is a ceramide. Other common groups bonded to the terminal oxygen atom include phosphocholine, yielding a sphingomyelin, and various sugar monomers or dimers, yielding cerebrosides and globosides, respectively. Cerebrosides and globosides are collectively known as glycosphingolipids.

Protein kinase B (PKB), also known as Akt, is the collective name of a set of three serine/threonine-specific protein kinases that play key roles in multiple cellular processes such as glucose metabolism, apoptosis, cell proliferation, transcription, and cell migration.
The exocyst is an octameric protein complex involved in vesicle trafficking, specifically the tethering and spatial targeting of post-Golgi vesicles to the plasma membrane prior to vesicle fusion. It is implicated in a number of cell processes, including exocytosis, cell migration, and growth.

The mammalian target of rapamycin (mTOR), also referred to as the mechanistic target of rapamycin, and sometimes called FK506-binding protein 12-rapamycin-associated protein 1 (FRAP1), is a kinase that in humans is encoded by the MTOR gene. mTOR is a member of the phosphatidylinositol 3-kinase-related kinase family of protein kinases.

Phosphoinositide phospholipase C is a family of eukaryotic intracellular enzymes that play an important role in signal transduction processes. These enzymes belong to a larger superfamily of Phospholipase C. Other families of phospholipase C enzymes have been identified in bacteria and trypanosomes. Phospholipases C are phosphodiesterases.
Phosphatidylinositol (3,4,5)-trisphosphate (PtdIns(3,4,5)P3), abbreviated PIP3, is the product of the class I phosphoinositide 3-kinases' (PI 3-kinases) phosphorylation of phosphatidylinositol (4,5)-bisphosphate (PIP2). It is a phospholipid that resides on the plasma membrane.

Phosphatidylinositol (3,4)-bisphosphate is a minor phospholipid component of cell membranes, yet an important second messenger. The generation of PtdIns(3,4)P2 at the plasma membrane activates a number of important cell signaling pathways.
In molecular biology, BAR domains are highly conserved protein dimerisation domains that occur in many proteins involved in membrane dynamics in a cell. The BAR domain is banana-shaped and binds to membrane via its concave face. It is capable of sensing membrane curvature by binding preferentially to curved membranes. BAR domains are named after three proteins that they are found in: Bin, Amphiphysin and Rvs.

WAS/WASL-interacting protein (WIP) is a protein that in humans is encoded by the WIPF1 gene.

PIKfyve, a FYVE finger-containing phosphoinositide kinase, is an enzyme that in humans is encoded by the PIKFYVE gene.
The oxysterol-binding protein (OSBP)-related proteins (ORPs) are a family of lipid transfer proteins (LTPs). Concretely, they constitute a family of sterol and phosphoinositide binding and transfer proteins in eukaryotes that are conserved from yeast to humans. They are lipid-binding proteins implicated in many cellular processes related with oxysterol, including signaling, vesicular trafficking, lipid metabolism, and nonvesicular sterol transport.
The Akt signaling pathway or PI3K-Akt signaling pathway is a signal transduction pathway that promotes survival and growth in response to extracellular signals. Key proteins involved are PI3K and Akt.

Autophagy-related protein 8 (Atg8) is a ubiquitin-like protein required for the formation of autophagosomal membranes. The transient conjugation of Atg8 to the autophagosomal membrane through a ubiquitin-like conjugation system is essential for autophagy in eukaryotes. Even though there are homologues in animals, this article mainly focuses on its role in lower eukaryotes such as Saccharomyces cerevisiae.
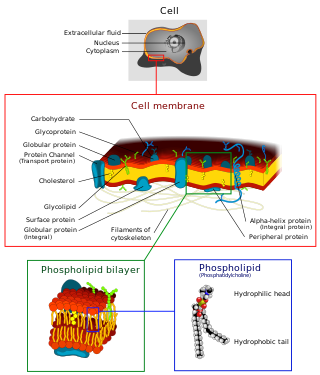
The cell membrane is a biological membrane that separates and protects the interior of a cell from the outside environment. The cell membrane consists of a lipid bilayer, made up of two layers of phospholipids with cholesterols interspersed between them, maintaining appropriate membrane fluidity at various temperatures. The membrane also contains membrane proteins, including integral proteins that span the membrane and serve as membrane transporters, and peripheral proteins that loosely attach to the outer (peripheral) side of the cell membrane, acting as enzymes to facilitate interaction with the cell's environment. Glycolipids embedded in the outer lipid layer serve a similar purpose. The cell membrane controls the movement of substances in and out of a cell, being selectively permeable to ions and organic molecules. In addition, cell membranes are involved in a variety of cellular processes such as cell adhesion, ion conductivity, and cell signalling and serve as the attachment surface for several extracellular structures, including the cell wall and the carbohydrate layer called the glycocalyx, as well as the intracellular network of protein fibers called the cytoskeleton. In the field of synthetic biology, cell membranes can be artificially reassembled.
mTOR Complex 2 (mTORC2) is an acutely rapamycin-insensitive protein complex formed by serine/threonine kinase mTOR that regulates cell proliferation and survival, cell migration and cytoskeletal remodeling. The complex itself is rather large, consisting of seven protein subunits. The catalytic mTOR subunit, DEP domain containing mTOR-interacting protein (DEPTOR), mammalian lethal with sec-13 protein 8, and TTI1/TEL2 complex are shared by both mTORC2 and mTORC1. Rapamycin-insensitive companion of mTOR (RICTOR), mammalian stress-activated protein kinase interacting protein 1 (mSIN1), and protein observed with rictor 1 and 2 (Protor1/2) can only be found in mTORC2. Rictor has been shown to be the scaffold protein for substrate binding to mTORC2.

The SNX8 is a sorting nexin protein involved in intracellular molecular traffic from the early endosomes to the TGN. It is suggested that it acts as an adaptor protein in events related to immune response and cholesterol regulation, for example. As a protein of the SNXs family, the SNX8 is formed of 465 aminoacids and presents a BAR-domain and a PX-domain which are very relevant in relation to its functions. Furthermore, SNX8 study is motivated by its medical significance in relation to diseases such as Alzheimer's Disease, cancer, neurodevelopmental malformations and to its role in fighting against viral infections.

David G. Drubin is an American biologist, academic, and researcher. He is a Distinguished Professor of Cell and Developmental Biology at the University of California, Berkeley where he holds the Ernette Comby Chair in Microbiology.
The arrestin family of proteins is subdivided into α-arrestins (also referred to as arrestin-related trafficking adaptors or arrestin-like yeast proteins in yeast or ARRDCs in mammals, β-arrestins and Vps26-like arrestins proteins. The α-Arrestins are an ancestral branch of the larger arrestin family of proteins and they are conserved across eukaryotes but are best characterized in the budding yeast Saccharomyces cerevisiae; to-date there are 6 α-arrestins identified in mammalian cells and 14 α-arrestins identified in the budding yeast Saccharomyces cerevisiae. The yeast α-arrestin family comprises Ldb19/Art1, Ecm21/Art2, Aly1/Art6, Aly2/Art3, Rod1/Art4, Rog3/Art7, Art5, Csr2/Art8, Rim8/Art9, Art10, Bul1, Bul2, Bul3 and Spo23. The best characterized α-arrestin function to date is their endocytic regulation of plasma membrane proteins, including G-protein coupled receptors and nutrient transporters. α-Arrestins control endocytosis of these membrane proteins in response to cellular stressors, including nutrient or metal ion excess.
References
- 1 2 Walther, Tobias C.; Brickner, Jason H.; Aguilar, Pablo S.; Bernales, Sebastián; Pantoja, Carlos; Walter, Peter (2006). "Eisosomes mark static sites of endocytosis". Nature. 439 (7079): 998–1003. Bibcode:2006Natur.439..998W. doi:10.1038/nature04472. PMID 16496001.
- 1 2 Deng, C.; Xiong, X.; Krutchinsky, A. N. (2009). "Unifying Fluorescence Microscopy and Mass Spectrometry for Studying Protein Complexes in Cells". Molecular & Cellular Proteomics. 8 (6): 1413–23. doi: 10.1074/mcp.M800397-MCP200 . PMC 2690482 . PMID 19269952.
- ↑ Seger S and Philippsen P (2010), Personal Communication to SGD
- ↑ "Saccharomyces Genome Database".
- 1 2 Olivera-Couto, A.; Grana, M.; Harispe, L.; Aguilar, P. S. (2011). "The eisosome core is composed of BAR domain proteins". Molecular Biology of the Cell. 22 (13): 2360–72. doi:10.1091/mbc.E10-12-1021. PMC 3128537 . PMID 21593205.
- 1 2 Moreira, K. E.; Walther, T. C.; Aguilar, P. S.; Walter, P. (2008). "Pil1 Controls Eisosome Biogenesis". Molecular Biology of the Cell. 20 (3): 809–18. doi:10.1091/mbc.E08-03-0313. PMC 2633383 . PMID 19037108.
- ↑ Kamble, Chitra; Jain, Sandhya; Murphy, Erin; Kim, Kyoungtae (2011). "Requirements of Slm proteins for proper eisosome organization, endocytic trafficking and recycling in the yeast Saccharomyces cerevisiae". Journal of Biosciences. 36 (1): 79–96. doi:10.1007/s12038-011-9018-0. PMID 21451250.
- ↑ Zhang, X; Lester, RL; Dickson, RC (May 21, 2004). "Pil1p and Lsp1p negatively regulate the 3-phosphoinositide-dependent protein kinase-like kinase Pkh1p and downstream signaling pathways Pkc1p and Ypk1p". The Journal of Biological Chemistry. 279 (21): 22030–8. doi: 10.1074/jbc.M400299200 . PMID 15016821.